
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
Prepare sequencing libraries from mRNA to get a clear view of the coding transcriptome with strand-specific information.
Assay time
Hands-on time
Input quantity
TruSeq Stranded mRNA offers a streamlined, cost-efficient, and scalable solution for coding transcriptome analysis. It is compatible with a wide range of samples.
Get precise measurement of mRNA strand orientation for detection of antisense transcription, enhanced transcript annotation, and increased alignment efficiency. High coverage uniformity enhances the discovery of features such as alternative transcripts, gene fusions, and allele-specific expression.
Stranded information identifies from which of the 2 DNA strands a given RNA transcript was derived. This information provides increased confidence in transcript annotation, particularly for nonhuman samples. Identifying strand origin increases the percentage of reads that align, reducing sequencing costs per sample.
The workflow enables robust interrogation of both standard and low-quality samples, and workflows compatible with a wide range of study designs.
Prepare libraries for Illumina sequencing in less than a day. Illumina Qualified Methods are available on a range of automation platforms through our partners.
| Assay time | ~10.5 hr |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Automation details | Explore available automation methods |
| Content specifications | Captures the coding transcriptome with strand information |
| Description | Gives researchers a clear, comprehensive view of the coding transcriptome with precise strand information. |
| Hands-on time | ~4.5 hr |
| Input quantity | 0.1 – 1 ug total RNA or 10 - 100 ng previously isolated mRNA (from species with polyA tails) |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Oligo-dT beads capture polyA tails |
| Method | mRNA sequencing |
| Multiplexing | 1–96 |
| Nucleic acid type | RNA |
| Specialized sample types | Not FFPE-compatible |
| Species category | Mammalian, Bovine, Mouse, Human, Rat |
| Species details | Works with high-quality RNA from any species with polyA tails |
| Strand specificity | Stranded |
| Technology | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
Illumina now offers modular product ordering to enable flexibility in your workflows. You may order any of the following library prep products with any of the following indexes.
TruSeq Stranded mRNA
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 550 System | RNA Profiling: 13–40 samples per run (based on 10 million reads per sample) |
2 x 75 bp |
| NovaSeq 6000 System | RNA Profiling (samples per run, dual flow cell): S1: 320, S2:384, S4:768. Limited by index combinations (dual). Based on 10 million reads. |
≤ 2 × 100 bp |
mRNA sequencing (mRNA-Seq) using NGS can identify both known and novel transcripts, and measure transcript abundance.
Detect cancer gene expression and transcriptome changes and identify novel cancer transcripts with RNA next-generation sequencing (RNA-Seq).
Learn how to profile gene expression changes for a deeper understanding of biology.
Researchers at the Icahn School of Medicine at Mount Sinai are studying mtDNA, piRNA, and TCR repertoire diversity to unlock the secrets of cancer, infertility, and autoimmune disease.
RNA-Seq and HLA typing are increasing the power and efficiency of a target discovery platform.
| TruSeq Stranded mRNA | Illumina Stranded mRNA Prep | Illumina RNA Prep with Enrichment | |
|---|---|---|---|
| Assay time | ~10.5 hr | 6.5 hr | < 9 hr |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) | Liquid handling robot(s) |
| Automation details | Explore available automation methods | Explore available automation methods | |
| Content specifications | Captures the coding transcriptome with strand information | Captures the coding transcriptome with strand information | Captures the coding transcriptome when used with Illumina Exome Panel |
| Description | Gives researchers a clear, comprehensive view of the coding transcriptome with precise strand information. | A simple, cost-effective solution for analysis of the coding transcriptome with precise strand information | A reproducible, economical solution enabling targeted transcript detection and discovery from a broad range of sample types and inputs including formalin-fixed, paraffin- embedded (FFPE) tissues and other low-quality samples |
| Hands-on time | ~4.5 hr | < 3 hr | < 2 hr |
| Input quantity | 0.1 – 1 ug total RNA or 10 - 100 ng previously isolated mRNA (from species with polyA tails) | 25-1000 ng standard-quality total RNA | 10 ng RNA; 20 ng FFPE RNA |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System, NovaSeq 6000 System, MiSeq i100 Plus System, MiSeq i100 System |
| Mechanism of action | Oligo-dT beads capture polyA tails | PolyA capture, ligation-based addition of adapters and indexes | Bead-linked transposome |
| Method | mRNA sequencing | mRNA sequencing | mRNA sequencing, Targeted RNA sequencing, Target enrichment |
| Multiplexing | 1–96 | Up to 384 Unique Dual Indexes (UDIs) | Up to 384 Unique Dual Indexes (UDIs) |
| Nucleic acid type | RNA | RNA | RNA |
| Specialized sample types | Not FFPE-compatible | Not FFPE-compatible, Low-input samples | Blood, Low-input samples, FFPE tissue |
| Species category | Mammalian, Bovine, Mouse, Human, Rat | Mammalian, Bovine, Mouse, Human, Rat | Human, Virus |
| Species details | Works with high-quality RNA from any species with polyA tails | Works with high-quality RNA from any species with polyA tails | |
| Strand specificity | Stranded | Stranded | Non-stranded |
| Variant class | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
Library Prep and Array Kit Selector
Determine the best library prep kit or array for your needs based on your starting material and method of interest.
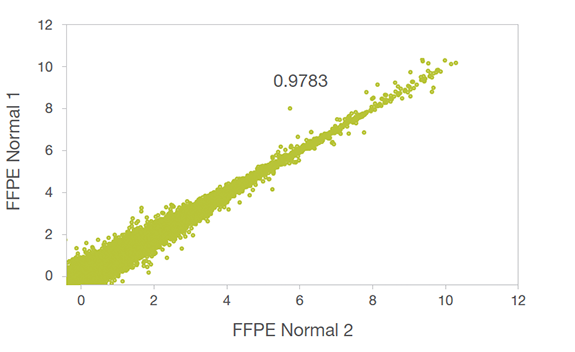

Technical Replicates
Technical replicates of FFPE tissue show high concordance, indicating robust library prep performance. Axes are log2(FPKM). R2 value is shown.
TruSeq® Stranded mRNA Library Prep (48 Samples)
20020594
Includes RNA purification beads for upfront polyA capture step, PCR master mix to transcript mRNA into cDNA, and reagents for preparing 48 samples. Purchase index adapters separately. Purfication beads are required post polyA capture step and are sold separately.
List Price:
Discounts:
TruSeq Stranded mRNA Library Prep (96 Samples)
20020595
Includes RNA purification beads for upfront polyA capture step, PCR master mix to transcript mRNA into cDNA, and reagents for preparing 48 samples. Purchase index adapters separately. Purfication Beads are required post polyA capture step and are sold separately.
List Price:
Discounts:
TruSeq RNA Single Indexes Set A (12 Indexes, 48 Samples)
20020492
Includes 12 of 24 indexes (Indexes: 2, 4, 5, 6, 7, 12, 13, 14, 15, 16, 18, 19) for preparing 48 individual RNA samples.
List Price:
Discounts:
TruSeq RNA Single Indexes Set B (12 Indexes, 48 Samples)
20020493
Includes 12 of 24 indexes (Indexes: 1, 3, 8, 9, 10, 11, 20, 21, 22, 23, 25, 27) for preparing 48 individual RNA samples.
List Price:
Discounts:
IDT for Illumina – TruSeq RNA UD Indexes v2 (96 Indexes, 96 Samples)
20040871
Includes 96, 8 bp indexes sufficient for labeling 96 samples and is compatible with TruSeq Stranded mRNA and TruSeq Stranded Total RNA Library Prep Kits. Version 2 contains improvements for 8 of the 192 index oligos in plate 20022371, with no changes to the remaining 184. Purchase library preps and probe panels separately.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
This product and its use are the subject of one or more issued and/or pending U.S. and foreign patent applications owned by Max Planck Gesellschaft, exclusively licensed to New England Biolabs, Inc. and sublicensed to Illumina, Inc. The purchase of this product from Illumina, Inc., its affiliates, or its authorized resellers and distributors conveys to the buyer the non-transferable right to use the purchased amount of the product and components of the product by the buyer (whether the buyer is an academic or for profit entity). The purchase of this product does not convey a license under any claims in the foregoing patents or patent applications direct to producing the product. The buyer cannot sell or otherwise transfer this product or its components to a third party or otherwise use the product for the following COMMERCIAL PURPOSES: (I) use of the product or its components in manufacturing; or (2) use of the product or its components for therapeutic or prophylactic purposes in human or animals.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.