NGS in CRISPR Genome Editing
CRISPR Genome Editing Technology
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) genome editing is a revolutionary method in which a programmable RNA targets a nuclease (eg, Cas9) to a specific location in the genome.1,2 The speed, simplicity, and precision with which CRISPR-Cas9 technology enables genetic elements to be mutated, silenced, induced, or replaced has resulted in its widespread adoption in the global research community.
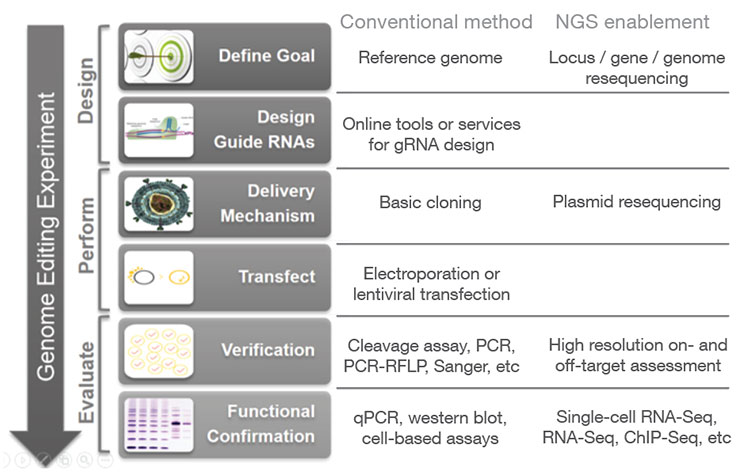
Next-generation sequencing (NGS) may be used at various stages of a genome editing workflow, from analyzing CRISPR off-target effects with whole-genome sequencing to confirming CRISPR knockouts and other edits with targeted sequencing. Follow-up studies can then be performed using applications such as methylation analysis and gene expression profiling with RNA sequencing, in order to assess the functional impact of a given gene edit.
Gene Editing Publication Review
See summaries of recent peer-reviewed gene editing research publications featuring Illumina NGS technology.
Read ReviewApplications of CRISPR Genome Editing
Applications of CRISPR-Cas9 technology have been identified in the fields of basic and clinical research, therapeutics, drug development, agriculture, and the environment. Clinical research has shown potential utilization for CRISPR in such diseases as sickle cell disease, cancer, AIDS, Huntington’s disease, Duchenne muscular dystrophy, and more.
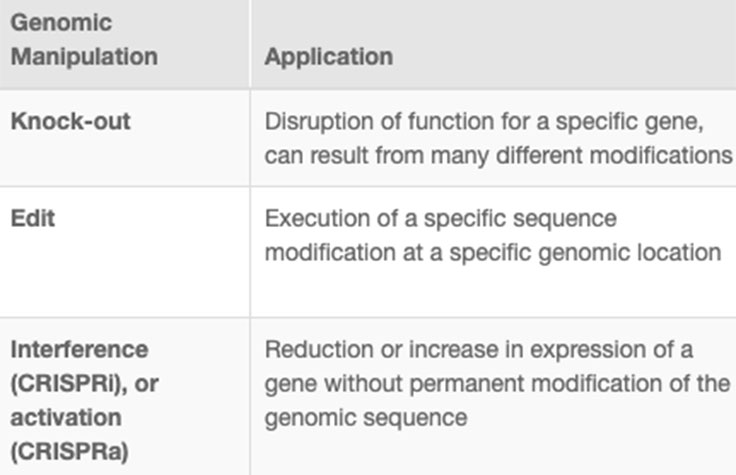
CRISPR genome editing allows researchers to create genetically modified cell lines and animal models with speed and precision. In addition to creating gene knockouts and gene knock-ins, researchers can use CRISPR technology to modulate gene expression via interference (CRISPRi) or activation (CRISPRa), without altering the genomic sequence. View the associated table to learn more.
Confirm CRISPR Knockouts and Other Edits
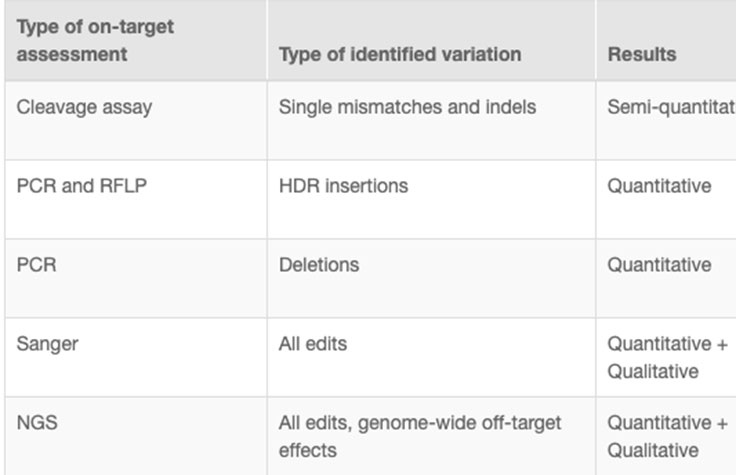
CRISPR genome editing experiments result in mixed cell populations, with only a small subset carrying the desired edit. Researchers need to determine which cells have the desired CRISPR knockout or targeted mutation. Current methods to evaluate edits involve cleavage assays, PCR, Sanger sequencing, and NGS. View the associated table for additional information.
NGS is the only assay that provides both qualitative and quantitative information at high resolution across the full range of modifications, meets the needs of any throughput, and can be used to monitor off-target effects.7 NGS-based targeted sequencing provides a cost-effective solution for confirming CRISPR-induced edits by focusing on regions targeted for modification.
Learn more about targeted sequencingCRISPR Off-Target Analysis with NGS
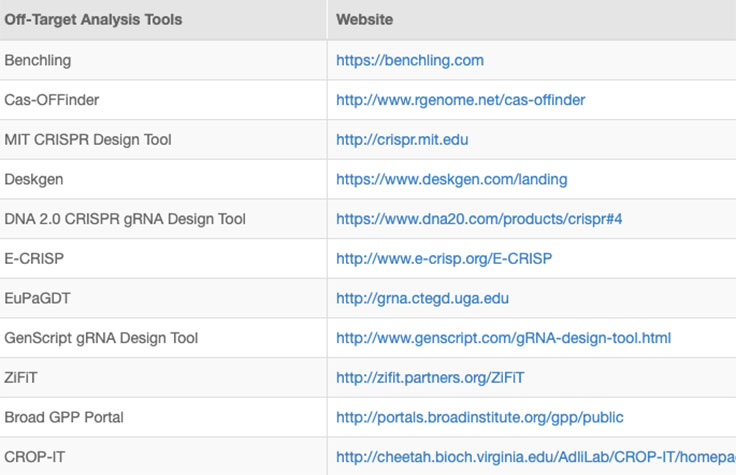
Successful implementation of CRISP/Cas9 technology should include strategies to identify and reduce off-target effects, or unintended modifications at sites other than the intended target. Computational methods to evaluate RNA specificity and predict off-target sites are commonly used during genome editing experiments.
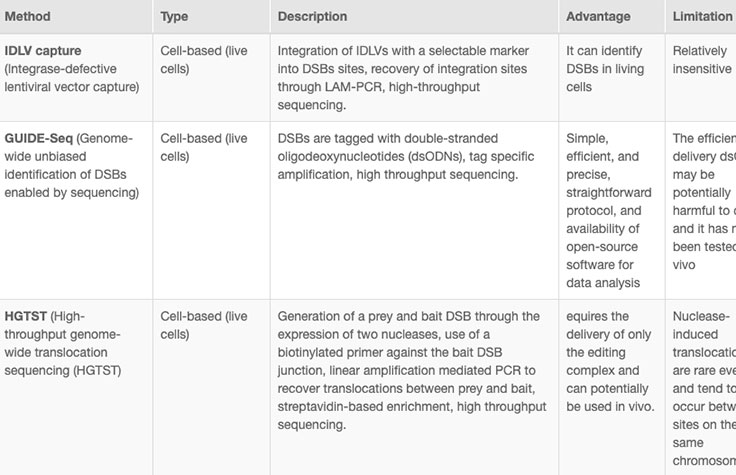
Online tools and web-based algorithms are publicly available, as shown in the "Off-Target Analysis Tools" table. However, genome-wide analyses such as NGS-based whole-genome sequencing (WGS) are often necessary to discover off-target sites that may escape prediction algorithms.8
Learn more about WGSFeatured CRISPR-Cas9 Genome Editing Content

Genome Editing and CRISPR-Cas9: Podcast Episode 32
Dr. Sam Sternberg of Columbia University discusses the biology and impact of CRISPR and gene editing.
Listen Now
CRISPR-Cas9: Genome Engineering Made Simple
The Illumina Scientific Affairs team summarizes key publications on applications of CRISPR-Cas9 technology.
View Video
Long Non-Coding RNAs and Cancer
Researchers discuss recent cancer-related lncRNA studies, from biomarker discovery to CRISPR- and siRNA-based approaches for silencing cancer-specific lncRNAs.
Read InterviewAssess the Functional Impact of CRISPR Genome Edits
Scientists can use a vast repertoire of sequencing methods to determine the impact of an edited sequence on the structure and function of genes. Some methods commonly used to study the functional effects of gene edits include:
Single-Cell RNA-Seq
Screen cell populations after CRISPR modification to determine the gene-regulatory impact of many genes in parallel in thousands of individual cells.
RNA Sequencing
Assess the impact of mutations on the transcriptome as a whole or on the expression of genes/gene families.
ChIP-Seq
Determine the impact of genome edits on DNA-protein binding.
Methylation Sequencing
Investigate the downstream impact of mutations on methylation status and chromatin remodeling.
Featured Products
More Uses of NGS in Genome Editing
In addition to high resolution on- and off-target assessment and functional analysis of CRISPR edits, NGS can be incorporated at additional stages of the CRISPR genome editing workflow.
During the initial design phase, resequencing of a locus or genome (for species that lack a reference genome) can aid in RNA selection. During the process of cloning CRISPR-Cas9/guide RNA constructs, resequencing of the resulting plasmids can provide rapid and high-confidence verification of the CRISPR delivery vectors, especially for high-throughput experiments with large plasmid libraries.
Francis deSouza on Genomics, CRISPR, and the Future
Francis deSouza, President and CEO of Illumina, Inc., took the stage at Aspen Ideas: Health in Colorado to educate the audience on the impact genomics is having across healthcare, and the urgency in bringing its benefits to more people, more quickly.
Read Article
Related Solutions
Agrigenomics

The simplicity and low cost of CRISPR-Cas9 technology can extend gene editing in crops from large commodity species to a wider variety of agriculturally important species.
Learn more about agricultural genomicsCancer Research

The speed and simplicity of CRISPR-Cas9 technology can facilitate development of cancer models and discovery of new immunotherapeutic targets and strategies.
Learn more about cancer researchComplex Disease Genomics

The precision of CRISPR-Cas9 genome editing technology can facilitate development of cell and animal models of human complex diseases to investigate disease pathology.
Learn more about complex disease researchCell and Molecular Biology

The speed, simplicity, and low cost of CRISPR-Cas9 gene editing has revolutionized cell and molecular biology in the development of gene knockouts and transgenic models.
Learn more about cellular and molecular biology researchAdditional Resources
NGS Technology
Ultra-high throughput, scalability, and speed enable biological understanding never before possible.
Publication Reviews
These summaries of peer-reviewed publications highlight how Illumina technology is furthering scientific research.
References
- Cong L, Ran F A, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823.
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-826.
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538-542.
- Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543-548.
- Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173-1183.
- Cheng AW, Wang H, Yang H, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163-1171.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281-2308.
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300-312.
- Gabriel R, Lombardo A, Arens A, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816-823.
- Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187-197.
- Chiarle R, Zhang Y, Frock RL, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107-119.
- Crosetto N, Mitra A, Silva MJ, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361-365.
- Kim D, Kim S, Kim S, Park J, Kim JS. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016;26:406-415.
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14:607-614.