Karyomapping
Get Faster Results More Efficiently with Karyomapping
Karyomapping is a technique used to test in vitro embryos for genetic mutations. It relies on the use of genetic markers within the genome to assess the likelihood of an in vitro embryo carrying a gene mutation.
Alternate methods for identifying single-gene disorders examine the inheritance of short tandem repeats (STRs) adjacent to the disorder’s specific loci. STR methods are expensive and time-consuming because they are developed for each test run.
Our karyomapping solution is a faster alternative to STR analysis.1 Using genome-wide linkage-based analysis from a single cell, karyomapping detects whether the gene region containing the mutation was transmitted to an embryo.

The Benefits of Karyomapping
The karyomapping technique can create a complete map of the in vitro embryo chromosomes, giving researchers valuable insight into the role that genomics plays in the inheritance of single-gene defects.
Karyomapping analysis uses genome-wide linkage to reveal the inheritance of genetic disease loci present in one or both parents. Instead of STRs flanking a gene of interest as markers, karyomapping uses single nucleotide polymorphisms (SNPs). A SNP is a DNA sequence variation occurring when a single nucleotide in the genome differs between individuals. They are easily identified using existing genome-wide SNP technologies. Karyomapping compares SNPs from an in vitro embryo to those of a reference to establish the likelihood of the embryo having inherited the mutated gene.
Karyomapping offers a number of advantages over STR analysis, including:
- SNPs are fairly evenly distributed over the entire genome.
- The HumanKaryomap-12 BeadChip provides a large number of data points, reducing the risk of missing a recombination event in the genomic region of interest.
- Karyomapping does not require custom design of probes for each inherited single-gene disorder, resulting in significant time savings.
- Karyomapping is amenable to automated analysis using software for consistent results.
A Comparison of STR Analysis and Karyomapping
| STR Analysis | Karyomapping | |
|---|---|---|
| Description | Uses DNA repeats of 2–6 base pairs adjacent to a specific locus to identify a defective gene |
Uses SNPs and genome-wide linkage data to inform the presence or absence of a defective gene |
| Markers | Multiallelic; measures variation in repeat length |
Biallelic; measures variation at a single base |
| Coverage | Limited to a single locus in each set of STR markers |
Able to screen multiple loci in parallel |
| Location | Requires knowledge of the location of the affected gene |
Requires knowledge of the location of the affected gene |
| Preparation Time | Typically 3–6 months to work up and validate multiple STR markers |
None; off-the-shelf solution |
| Workflow | Customized set of primers for each case |
Standard workflow for all studies |
| Linkage Analysis | Performed manually |
Automated data interpretation using BlueFuseMulti analysis software |
| Aneuploidy | Not available |
Not currently offered |
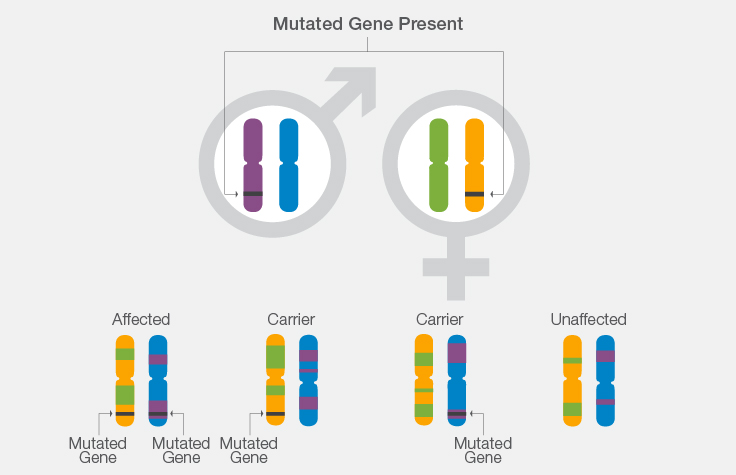
How Karyomapping Works: A Cystic Fibrosis Example
- If both parents carry a mutation for cystic fibrosis, there is a 1 in 4 chance that their offspring will have the disease.
- Using knowledge of CF carrier or disease status in the parents, scientists can look at the chromosome segment in which the gene lies to determine from which parental chromosome the gene originated.
- Scientists can then determine whether the chromosome segment inherited by each in vitro embryo contains the normal or mutated copy of the CF gene.
- This helps determine whether the in vitro embryo inherited a single mutation from either parent and will be an unaffected carrier, whether the in vitro embryo inherited a mutation from each parent and will be affected with CF, or whether the in vitro embryo did not inherit any mutations.
Karyomapping Products
Illumina offers a comprehensive karyomapping solution that includes:
NextSeq 550 System
A desktop sequencing system that provides the flexible power and simplicity you need to analyze whole genomes, exomes, and transcriptomes. Optimized sequencing reagents and clustering chemistry yield the data quality you’ve come to expect from Illumina.
BlueFuse Multi Software
It offers a clear, intuitive user interface; a common workflow; and scales for the needs of your lab. This widely implemented software is used in the majority of laboratories performing PGS.
References
- American Society for Reproductive Medicine. Frequently asked questions about infertility. ReproductiveFacts. Org Website Accessed April 4, 2016.