Pan-Cancer Analysis
What is Pan-Cancer Analysis?
Although all cancers are molecularly distinct, many share common driver mutations.1 Pan-cancer analysis involves assessing frequently mutated genes and other genomic abnormalities common to many different cancers, regardless of tumor origin. Using next-generation sequencing (NGS), pan-tumor projects such as The Cancer Genome Atlas2 have made significant contributions to our understanding of DNA and RNA variants across many cancer types.
Pan-cancer profiling with NGS panels enables researchers to comprehensively analyze a defined set of gene variants and aberrations associated with many common cancers, including both solid tumors and hematologic malignancies.
Pan-Cancer Profiling with Targeted NGS Panels
Targeted sequencing offers a simple, cost-effective approach for pan-cancer profiling studies. Expert-defined pan-cancer panels eliminate the need to isolate gene targets of NGS separately, and multiplexing enables many samples to be run at the same time regardless of tumor type. Multiplexing enables pan-cancer RNA studies to be run in parallel with DNA studies for a comprehensive tumor profile.
Rapid NGS Workflows
NGS-based pan-cancer analysis follows simple workflows that can be easily scaled to large numbers of samples. Pan-cancer analysis allows the inclusion of markers from various cancer types in a single assay, positively impacting workflows and turnaround times.
Featured Pan-Cancer Articles
A New View of Cancer Pathways in Pediatric Leukemia
Researchers use a targeted RNA sequencing pan-cancer panel to understand the role of fusion genes in pediatric leukemia.
Read Interview
NGS Panels Demonstrate Value in Brain Tumor Studies
Researchers describe studies to identify diffuse glioma genetic markers, and discuss the benefits of a pan-cancer panel.
Read Interview
The Complex World of Pan-Cancer Markers
Learn about promising pan-cancer biomarkers and efforts to recharacterize cancers as mutation-based rather than organ-based.
Read ArticlePan-Cancer Analysis Products
Our comprehensive portfolio of NGS-based pan-cancer panels and library preparation kits enables cancer researchers to run DNA and RNA assays simultaneously for a broad view of tumor mechanisms.
Illumina sequencers deliver industry-leading accuracy, generating ~90% of the world’s sequencing data.* Efficient reporting capabilities enable researchers to annotate, classify, and communicate significant findings in a standard format that facilitates data analyses and trending.
Click on the below to view products for each workflow step.
Offers comprehensive coverage of pan-cancer content for clinical research. Assesses 523 cancer-relevant genes, as well as tumor mutational burden and microsatellite instability.
TruSight RNA Pan-CancerResearch panel targeting 1385 oncology genes for gene expression, variant and fusion detection in all RNA sample types including FFPE.
AmpliSeq for Illumina Cancer HotSpot Panel v2Targeted research panel investigating hotspot regions of 50 genes with known associations to cancer.
A gene fusion detection panel targeting fusion associated genes in many cancer types with the ability to detect known and novel fusion gene partners.
TruSight Oncology UMI ReagentsThe TruSight Oncology UMI Reagents and UMI Error Correction App reduce error rates in samples to ≤0.007%, enabling the detection of low frequency variants.
Simplest and most affordable solution for low-throughput targeted sequencing for pan-cancer profiling.
MiSeq SystemDesktop sequencer featuring a simple NGS workflow, integrated data analysis software, and unmatched accuracy.
FDA-cleared in vitro diagnostic NGS system that also operates in research mode to generate accurate, reliable data.
NextSeq 550 SystemFlexible desktop sequencers that support multiple applications, from targeted profiling to whole-genome sequencing.
The Illumina genomics computing environment for NGS data analysis and management.
MiSeq ReporterEasy-to-use software for analysis and variant calling on the MiSeq System.
BaseSpace Variant InterpreterA powerful analysis and reporting tool that provides biological insight into genomic variant data.
Easy-to-use software for automated on-instrument data analysis.
Illumina DRAGEN Bio-IT PlatformProvides accurate, ultra-rapid secondary analysis of sequencing data, including somatic datasets.
NGS Workflow Finder
Take the guesswork out of your next workflow. The NGS Workflow Finder provides personalized solution recommendations and resources so you can sequence with confidence.
Find your NGS workflow todayLearn More
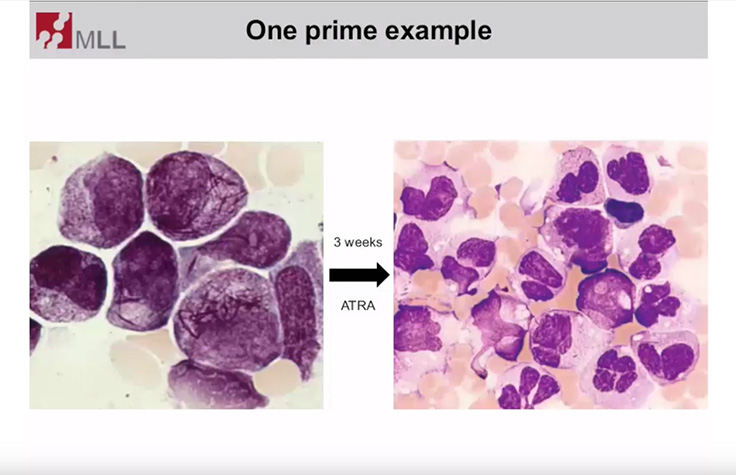
Targeted Sequencing for Hematologic Disorder Research
Dr. Torsten Haferlach, Head of Munich Leukemia Laboratory, discusses molecular assessments of hematologic disorders.
Nature Journal Focus: Pan-Cancer Analysis
This initiative examines similarities between genomic alterations across cancer types profiled by The Cancer Genome Atlas consortium.
Interested in receiving newsletters, case studies, and information on cancer genomics?
Sign UpReferences
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127-1133.
- Nature TCGA | TCGA Pan-Cancer Analysis (www.nature.com/tcga)
* Data calculations on file. Illumina, Inc. 2015.