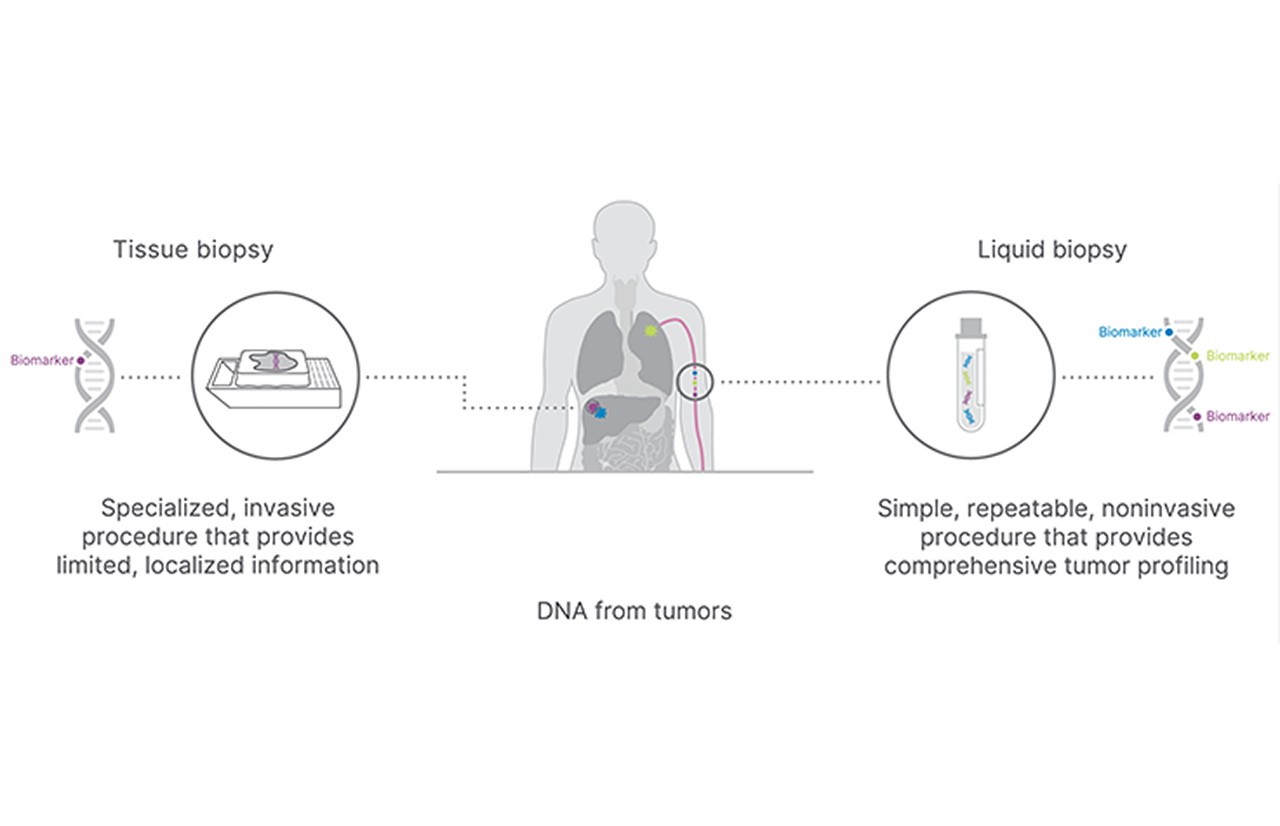
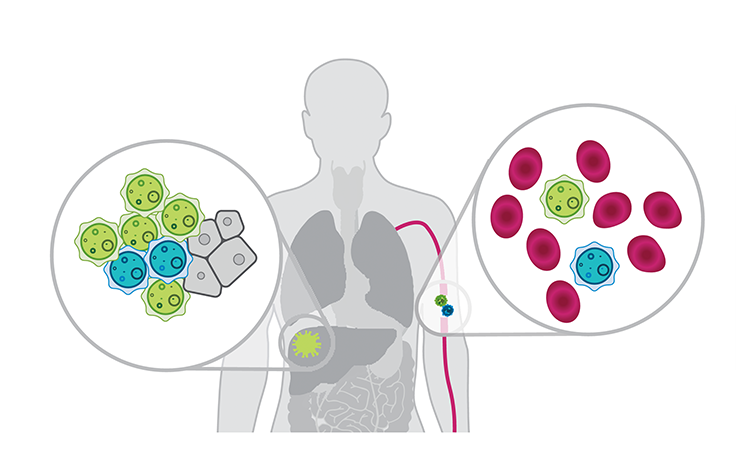
cfRNAs can be released from multiple cell types present in the bloodstream as well as from cancer cells. These ctRNAs are potential biomarkers that can be assayed by liquid biopsy to identify specific cancers, detect cancer initiation, reveal tissue-of-origin, elucidate molecular mechanisms of disease, monitor therapeutic response, and more.13
Unlike DNA, which is identical in every cell/tissue (except for genetic variants), RNA is dynamically and differentially expressed between cell types and tissues. This enables ctRNAs to be used to detect cancer and potentially localize it in the body. The diverse nature of RNA expression may also enable use of ctRNAs to determine and classify cancer subtypes early in disease, which is important given the wide range in progression, treatment options, and prognosis between cancers, even those associated with the same organ or tissue.14