RNA Sequencing Data Analysis Solutions
User-friendly RNA-Seq data analysis
Accessible software tools designed for biologists
Intuitive Analysis of RNA-Seq Data
Once in the domain of bioinformatics experts, RNA sequencing (RNA-Seq) data analysis is now more accessible than ever. Illumina offers push-button RNA-Seq software solutions packaged in intuitive user interfaces designed for biologists.
These user-friendly tools support a broad range of next-generation sequencing (NGS) studies, from gene expression analysis to total RNA expression profiling and more.
Benefits of RNA-Seq Data Analysis with Illumina Tools
RNA-Seq data can be quickly and securely transferred, stored, and analyzed in Illumina Connected Analytics or BaseSpace Sequence Hub, the Illumina cloud computing platforms. Both platforms offer in-cloud access to DRAGEN secondary analysis for accurate, ultra-rapid analysis of RNA-Seq and other NGS data.
DRAGEN uses lossless genomic data compression to reduce the data storage footprint by as much as 5x, all while preserving data integrity. Illumina Connected Multiomics offers user-friendly analysis and visualization of RNA-Seq as well as multiomics data, and accommodates input files from DRAGEN secondary analysis or select third-party platforms for maximum flexibility. Learn more about:
Our RNA-Seq analysis tools are:
- Accessible to any researcher, regardless of bioinformatics experience
- An expert-preferred suite of RNA-Seq software tools, developed or optimized by Illumina or from a growing ecosystem of third-party app providers
- Designed to support common transcriptome studies, from gene expression quantification to detection of novel transcripts, coding single nucleotide polymorphisms (cSNPs), gene fusions, and more
- Suitable for human, mouse, and rat RNA-Seq analysis (certain apps support additional species)
- Compatible with all Illumina sequencing systems

Core Lab Uses BaseSpace to Analyze mRNA-Seq Data
The BRC-Seq core sequencing facility at the University of British Columbia uses BaseSpace Sequence Hub to analyze and share data. Their studies frequently involve analysis of mRNA sequencing data to assess the impact of different experimental treatments on the transcriptome. The BaseSpace workflow helps BRC-Seq track samples and deliver high-quality sequencing data to its customers.
Read InterviewFeatured RNA-Seq Analysis Software
Based on frequently cited RNA-Seq data analysis pipelines, these software tools and apps support a broad range of transcriptome data analysis needs.
| Method | Featured Tool |
|---|---|
| mRNA Sequencing | |
| Targeted RNA Sequencing | |
| Small RNA Sequencing | |
| Infectious Disease Sequencing |
How to Analyze RNA-Seq Data with BaseSpace Apps
Walk through a typical BaseSpace Sequence Hub RNA sequencing data analysis workflow, from choosing your sequencing runs through selection of analysis parameters and analysis of results.
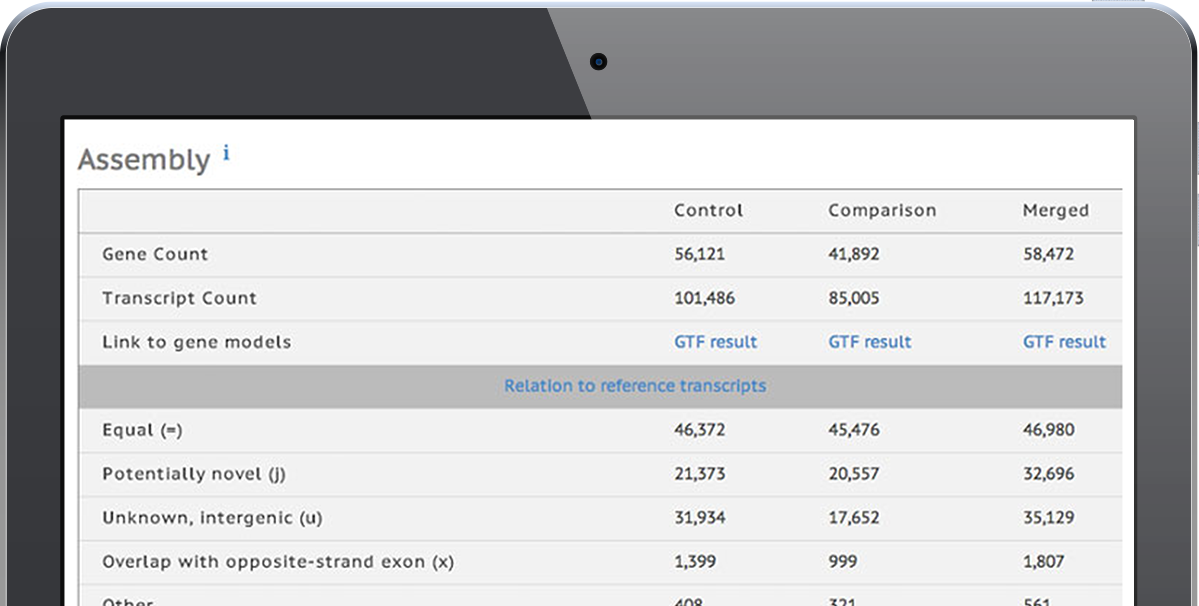
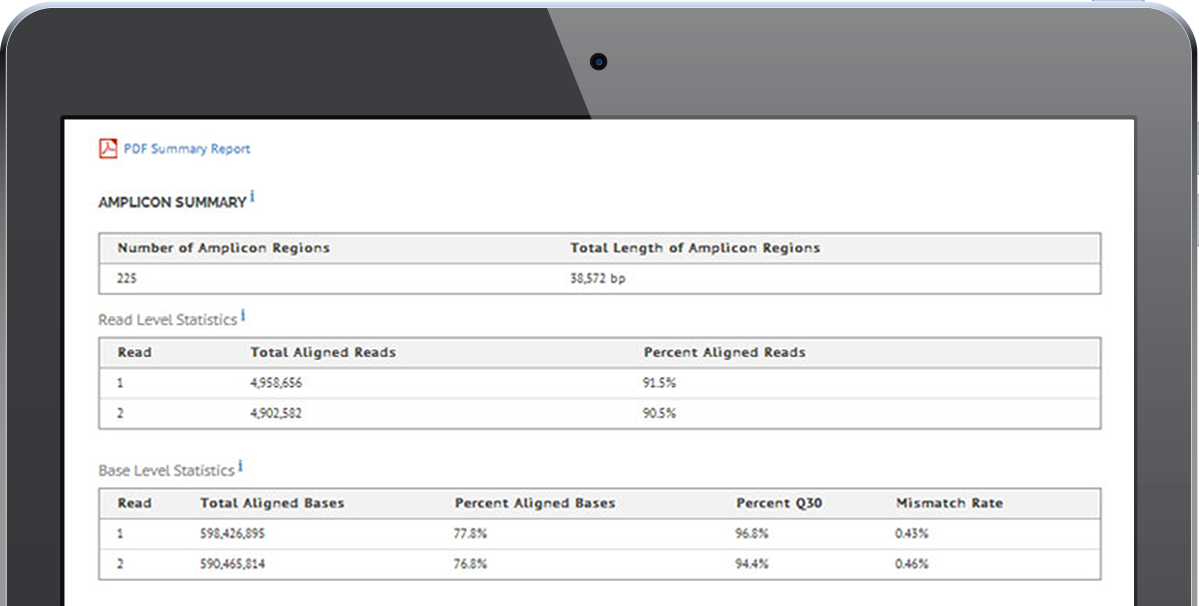
View examples of the easily interpretable tables and graphs that the software generates, including descriptions of differential expression, gene fusion detection, variant calling, and other reports that can be incorporated into a manuscript.
Read Technical NoteInterpretation of RNA-Seq Data
After data analysis, results can be transferred easily to Correlation Engine for functional annotation, to understand the biological effects of gene expression changes. This omics research database and suite of tools contains data sets from thousands of public studies that inform biological interpretation. The information is curated and normalized, giving the data additional power to help researchers define associations by tissue, disease, compound, and/or genetic perturbation.
Common applications of Correlation Engine include connecting differential gene expression data from RNA-Seq experiments with disease associations, and visualizing correlated genes and microRNA targets.
Explore Correlation EngineProviding Biological Context
Learn how Correlation Engine helps University of Pittsburgh researchers understand gene upregulation and downregulation, gene function, drug activity, and other mechanisms, and contextualize their results for publications and grants.
Read ArticleRNA-Seq Data Examples for Common Workflows
View Sample Data Sets
See sample data sets for various RNA-Seq methods in BaseSpace Sequence Hub, or test BaseSpace Apps and evaluate the results interactively.
Note that a customer login is required to access BaseSpace Sequence Hub and view specific data sets.
Multiomics Profiling
Combine RNA-Seq data with data from other modalities such as epigenetics and proteomics, to measure gene expression, gene regulation, and protein levels.
Learn More About MultiomicsRelated Content
How to Streamline Bulk RNA-Seq Analysis
Learn how to perform each data analysis step, from sequence read alignment to QC, read quantification, interpretation of differential gene expression, and more.
Lossless Genomic Data Compression
Lossless genomic data compression technology reduces the data storage footprint by as much as five times, and decreases data storage and transfer costs.
NextSeq 1000 and NextSeq 2000 System Applications
These benchtop sequencing systems fuel a broad range of applications, from mRNA-Seq to single-cell RNA-Seq, target enrichment, and more.
Interested in receiving newsletters, case studies, and information on genomic analysis techniques?
Enter your email address.