Cancer Biomarker Detection

Identify actionable cancer biomarkers
The number of cancer biomarkers with associated targeted therapies continues to increase. We are at a place now with NGS-based comprehensive genomic profiling where one biopsy, one test, and one report can lead to improved outcomes for cancer patients, thereby advancing precision medicine and improving patient health and wellness.1-2
There are several key considerations when evaluating a cancer biomarker test to aid with therapy selection.
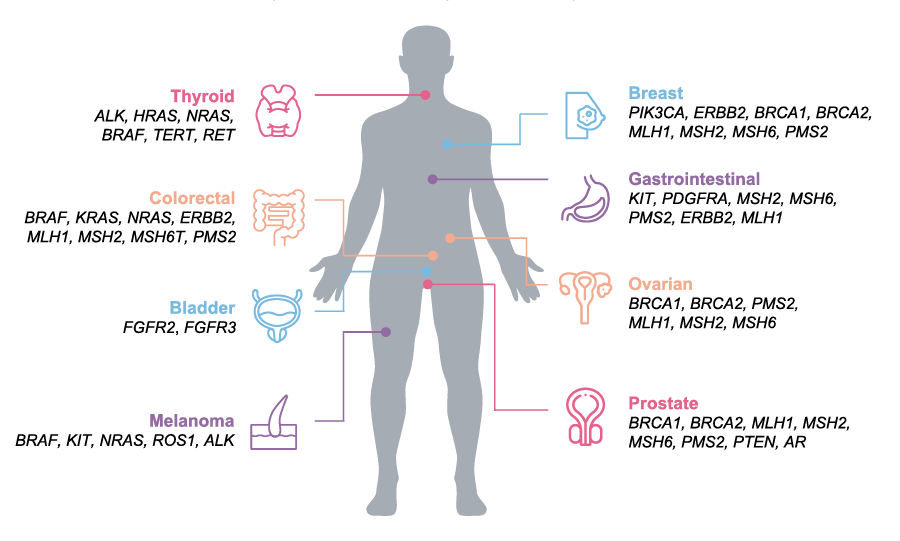
Biomarkers in US guidelines and drug labels for highly prevalent tumors 1,3
Maximize biomarker data from one biopsy
Conventional testing with techniques like FISH, PCR and IHC are insufficient to assess all relevant cancer biomarkers and preserve precious tissue samples. Individual sequential biomarker tests are common; however, they have some disadvantages. First, these tests require a significant amount of biopsy sample that’s not always accessible. Second, these single gene tests have limited content and may miss the opportunity to identify a positive biomarker.
A next-generation sequencing (NGS)-based biomarker test can analyze hundreds of clinically actionable cancer biomarkers simultaneously. This additional data provides more opportunity to match patients with appropriate molecular treatment regime. A single NGS-based biomarker test can replace multiple single-gene tests or small hotspot panels.4-7
If tissue biopsies are unavailable, comprehensive genomic profiling (CGP) from liquid biopsy may provide helpful information about a tumor's genomic make-up. This can decrease the need to rebiopsy and save precious time in the care continuum.8
Examples of key cancer biomarkers
Immunotherapy biomarkers
In addition to genes specifically involved in immune pathways, associated biomarkers have emerged that rely upon assessment of numerous genomic loci, such as tumor mutational burden (TMB) and microsatellite instability (MSI).
Gene fusions
Associated with the increasing number of discovered fusion events is the number of clinically actionable cancer biomarkers. Examples of clinically actionable gene fusions include NTRK and RET gene fusions.
Learn more about:
Homologous recombination deficiency (HRD)
HRD (homologous recombination deficiency) is a phenotype used to describe loss of function in the HRR (homologous recombination repair) pathway. When this occurs, cells are unable to repair double-stranded DNA breaks, leading to tumorigenesis.
HRD can be measured by "cause," such as a BRCA mutation or by "consequence" in the form of genomic scarring. NGS technology can assess both causal genes and genomic scarring.
SNVs, CNVs, & indels
There are multiple types of mutations that are associated with cancer-related genes, such as single-nucleotide variants (SNVs), insertions and deletions (indels), and copy-number variants (CNVs).
Traditionally these specific biomarkers have been analyzed with single-gene tests, but as the number of biomarkers increases for each cancer type, more labs have turned to NGS methods that can analyze numerous genes and variant types in a single assay.
CGP for cancer biomarker detection
Comprehensive genomic profiling (CGP) consolidates hundreds of cancer-related biomarkers into a single assay, eliminating the need for sequential testing. CGP can detect biomarkers at nucleotide-level resolution and typically comprises all major genomic variant classes (SNVs, indels, CNVs, fusions, and splice variants), as well as large genomic signatures (TMB and MSI), maximizing the ability to find clinically actionable alterations.
Learn more about CGPImproving patient outcomes with CGP
Maximize identification of molecularly matched therapies.
Cancer biomarkers for CGP test inclusion
CGP provides tumor-agnostic testing for hundreds of relevant cancer biomarkers, per guidelines, in a single assay, potentially offering significant savings in sample, time, and cost.
| Pan-Cancer Biomarkers NTRK1 NTRK2 NTRK3 MSI TMB RET BRAF | ||||||||
|---|---|---|---|---|---|---|---|---|
| Lung | Melanoma | Colon | Ovarian | Breast | Gastric | Bladder | Sarcoma | |
| AKT1 | BRAF | AKT1 | BRAF | AKT1 | BRAF | MSH5 | ALK | |
| ALK | CTNNB1 | BRAF | BRCA1 | AR | KIT | PMS2 | APC | |
| BRAF | GNA11 | HRAS | BRCA2 | BRCA1 | KRAS | TSC1 | BRAF | |
| DDR2 | GNAQ | KRAS | KRAS | BRCA2 | MET | CDK4 | ||
| EGFR | KIT | MET | PDGFRA | ERBB2 | MLH1 | CTNNB1 | ||
| ERBB2 | MAP2K1 | MLH1 | FOXL2 | FGFR1 | PDGFRA | ETV6 | ||
| FGFR1 | NF1 | MSH2 | TP53 | FGFR2 | TP53 | EWSR1 | ||
| FGFR3 | NRAS | MSH6 | PIK3CA | FOXO1 | ||||
| KRAS | PDGFRA | NRAS | PTEN | GLI1 | ||||
| MAP2K1 | PIK3CA | PIK3CA | KJT | |||||
| MET | PTEN | PMS2 | MDM2 | |||||
| NRAS | TP53 | PTEN | MYOD1 | |||||
| PIK3CA | SMAD4 | NAB2 | ||||||
| PTEN | TP53 | NF1 | ||||||
| RET | PAX3 | |||||||
| TP53 | PAX7 | |||||||
| PDGFRA | ||||||||
| PDGFRB | ||||||||
| SDHB | ||||||||
| SDHC | ||||||||
| SMARCB1 | ||||||||
| TFE3 | ||||||||
| WT1 | ||||||||
The genes shown here are not an exhaustive list.
Featured webinars
Weill Cornell pathology lab implements CGP
These walk-away automation methods for Illumina kits eliminate the need to develop your own method and can help you reduce hands-on time and minimize errors.
Emerging biomarkers in thyroid cancer and NSCLC
Dr. Sandip Patel of UCSD provides an overview of biomarker-guided therapies for thyroid cancer and non-small cell lung cancer (NSCLC) and discusses key emerging biomarkers for these indications along with their supporting evidence.
Broad oncology biomarker detection with CGP
Phil Febbo, MD discusses the benefits of CGP and how it’s becoming a new standard of care in oncology.
Scientists discuss cancer biomarker findings
Identifying biomarkers for response and resistance to anti-cancer immunotherapy
Dr Eliezer Van Allen highlights how NGS can identify biomarkers of response and resistance to immunotherapy, and can determine how cancers develop resistance to immune checkpoint blockade.
Accurate identification of cancer biomarkers
Scientists discuss the advantages of comprehensive NGS-based panels for biomarker discovery.
NGS proves invaluable for biomarker discovery
Phil Febbo, MD discusses the benefits of CGP and how it’s becoming a new standard of care in oncology.
Interested in receiving newsletters, case studies, and information on oncology?
Related content
Tumor mutational burden
NGS methods enable efficient assessment of tumor mutational burden and identification of neoantigens.
ctDNA as a noninvasive cancer biomarker
NGS methods enable efficient assessment of tumor mutational burden and identification of neoantigens.
Cancer epigenetics
NGS and microarray technologies can detect altered DNA methylation patterns and other epigenetic modifications that regulate cancer progression.
References
- US Food & Drug Administration. Hematology/Oncology (Cancer) Apporvals & Safety Notifications. US FDA website. Updated February 9, 2021. Accessed December 1, 2020.
- Singal G, Miller PG, Agarwala V, et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA. 2019 Apr 9;321(14):1391-1399. doi: 10.1001/jama.2019.3241.
- PierianDx. Next Generation Knowledge. PierianDx - Genomic Knowledgebase for Clinical Next Generation Knowledge. Accessed March 1, 2020.
- Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020 Jan 14;12(1):8. doi: 10.1186/s13073-019-0703-1.
- Reitsma M, Fox J, Borre PV, et al. Effect of a Collaboration Between a Health Plan, Oncology Practice, and Comprehensive Genomic Profiling Company from the Payer Perspective. J Manag Care Spec Pharm. 2019 May;25(5):601-611. doi: 10.18553/jmcp.2019.18309.
- Kopetz S, Mills Shaw KR, Lee JJ, et al. Use of a Targeted Exome Next-Generation Sequencing Panel Offers Therapeutic Opportunity and Clinical Benefit in a Subset of Patients With Advanced Cancers. JCO Precis Oncol. 2019 Mar 8;3:PO.18.00213. doi: 10.1200/PO.18.00213.
- Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015 Aug 15;21(16):3631-9. doi: 10.1158/1078-0432.CCR-14-2683.
- Rolfo C, Mack P, Scagliotti G, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the international association for the study of lung cancer. J Thorac Oncol. 2021 Oct;16, (10):1647-62. doi: 10.1016.