The Changing Landscape of NGS Implementation in Molecular Diagnostic Laboratories
Introduction
Next-generation sequencing (NGS) technology has accelerated the discovery of variants associated with disease in humans, particularly mutations involved in cancer. Some of these research discoveries became the basis for companion diagnostics and laboratory developed tests (LDT) now in use in molecular diagnostic laboratories worldwide. These laboratories are now faced with an evolving landscape of regulations and reimbursement strategies for NGS oncology testing.
At a recent Illumina User Group Meeting in Singapore, key opinion leaders from around the world took part in a panel discussion about the impact of NGS testing in oncology, now and in the future. Panelists included: Lawrence Jennings, MD, PhD, Northwestern University Feinberg School of Medicine; Simon Patton, PhD, European Molecular Genetics Quality Network; Min-Han Tan, MBBS, PhD, Lucence Diagnostics; and Benedict Yan, MBBS, National University Hospital Singapore. The audience participated in the panel discussion through a series of polling questions that highlighted various issues from new government testing standards to NGS testing frequency.1 Here we capture some of their insights into the changing landscape and headwinds facing NGS implementation in molecular diagnostic laboratories throughout the world.

From left to right: Lawrence Jennings, MD, PhD, Attending Pathologist, Director, HLA and Molecular Diagnostic Laboratory, Northwestern University Feinberg School of Medicine; Simon Patton, PhD, Director, European Molecular Genetics Quality Network (EMQN); Min-Han Tan, MBBS, PhD, Founder and CEO, Lucence Diagnostics; Benedict Yan, MBBS, Pathologist, Head, Molecular Diagnostics Centre, National University Hospital.
Q: How quickly do you see molecular genetics laboratories replacing single-gene tests with multigene NGS panels?
Lawrence Jennings (LJ): From the survey, most respondents believe they'll be shifting to multigene NGS panels in the future. Laboratories move from single-analyte testing to multianalyte NGS panel testing for two reasons. First, they know they’ll ultimately need to perform multianalyte testing on a single sample. The second reason is that the workflow for NGS panel testing is simpler than running multiple single-analyte tests.
Simon Patton (SP): We need to keep in mind that NGS is a technique. It’s really the clinical question being asked that’s important. If it’s not relevant to perform an NGS panel to answer the clinical question, why do it? If you can perform a test for three defined, recurrent mutations that are clearly present in that disorder, then there is no reason to use a broader NGS panel. It’s about using the right tool, or the right technique, to provide the right answer.
LJ: It also becomes an issue of what to report. From a clinical utility standpoint, laboratory clinicians might recognize the clinical relevance of analyzing a sample for BRAF, EGFR, NRAS, and KRAS. Obviously, it’s easier to analyze for them simultaneously. As we saw in one of the polling results (Figure 1), oncologists are requesting larger panels. If a lab runs a targeted 50-gene panel or a comprehensive 170-gene panel, the question becomes which subset of genes will be included in the report? Do they report on 50 genes or the smaller subset of KRAS, NRAS, BRAF, and EGFR?
Min-Han Tan (MHT): We operate across several countries and an important thing to understand is the differences in reimbursement in Asia, Europe, and America. In Asia, most of the payment is out of pocket for expensive diagnostics and medications. As a result, sequential testing remains important in many parts of Asia.
However, I agree that the tool must fit the clinical scenario. Sequential testing still makes sense in certain situations. NGS panels provide a clear advantage when biopsy tissue is hard to obtain or of poor quality. It is always about what provides the greatest clinical benefit and when.
Q: Polling respondents chose variant of unknown significance (VUS), cost, result accuracy, and result interpretation as their areas of concern surrounding cancer panel testing (Figure 2). Do you agree?
MHT: It depends on whether the cancer panel is for germline or somatic testing. For germline testing, one of the major issues is VUSs, which can be approached differently depending on the background skill and training of the clinician. For somatic testing, VUSs are less of an immediate concern because the majority of actionable mutations are quite clear.
LJ: Results interpretation can be an issue, especially if there are surprise variants. The discussion turns to ‘what is the course of action now that we know this variant is present?’
SP: In my proficiency testing (PT) schemes, I’ve found that accuracy is an issue. We see many errors, particularly in oncology and in new areas of test development and implementation. Our average genotyping error rate is just under 3%. When many of the new oncology tests were implemented, the error rates were about 25%. So, one in four test results were wrong. What’s alarming is that these were external quality assurance (EQA) scheme results where the lab knew that this was an EQA sample. What’s the true underlying error in a clinical scenario? It’s less than 25%, but it’s higher than you might expect.
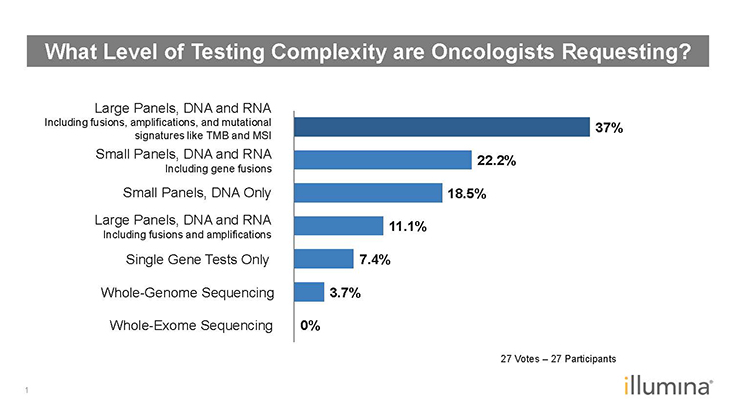
Figure 1: Poll Results–What Level of Testing Complexity are Oncologists Requesting?1
Q: How significant of an impact does the cost-per-reportable result have when deciding which tests to run?
MHT: The cost of NGS panels depends on the health care system and the reimbursement approach. In some US health care systems, NGS panels are covered. In Asia, most NGS panels are out-of-pocket expenses. In lung cancer, an NGS panel could reduce cost by reducing the overall time to answer or that liquid biopsies spare a patient procedure and thus reduce costs. As improved NGS panel validation occurs over time, and as the FDA and CFDAs review more submissions, access to NGS panels will increase given the clear clinical benefits.
Ben Yan (BY): Cost is definitely a concern for us. The recent US dollar increase has impacted our reagent costs in Singapore, increasing the cost per patient by $50 SGD.
LJ: From my perspective, the value is far more important. The costs are largely determined by the vendors who set reagent costs depending on what the market will allow. This is a reflection of the value the test offers and, more significantly, the reimbursement and costs of competitors’ reagents. Given two tests of equal value, cost becomes a factor. Other factors such as platform and standardization of workflow also come into play.
Q: Attendees also answered a multi-option question about when cancer patients should have their cancer samples sequenced–at diagnosis, at relapse, when nontargeted therapy options are exhausted, or not at all (Figure 3). It was evenly split between diagnosis, relapse, and when nontargeted therapy options are exhausted. Do you agree?
LJ: For solid tumors, I think it makes sense to perform sequencing in all three instances, certainly for diagnosis and relapse.
BY: For hematological cancers, it would be a good idea to sequence after the leukemia has been eradicated, but before the patient relapses. It’s possible that sequencing might show what mutations are still present.
LJ: We have clinicians who will order NGS panels for bone-marrow testing post-transplant, or when a patient is in remission. Most NGS panels are designed for diagnosis, and their limit of detection is 5%. For post-transplant and remission testing, you need a panel designed specifically to target minimal residual disease (MRD)-associated variants. Most tests that are used for diagnosis are not MRD tests. It’s important to realize that for MRD testing the limit of detection of the panel needs to be 0.1% or better.
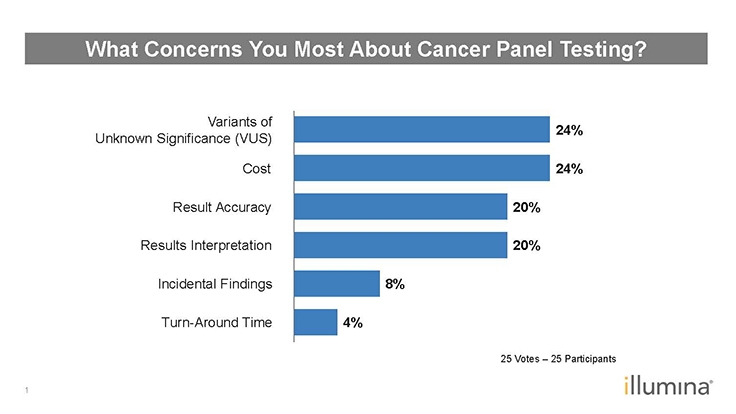
Figure 2: Poll Results–What Concerns You Most About Cancer Panel Testing?1
Q: Currently, tumor mutation burden (TMB) testing isn’t fully predictive, with different TMB tests providing different results. What is the EQA view concerning TMB testing?
SP: We plan to develop an EQA for TMB. Because of the reasons you’ve mentioned, we’re conducting a survey to understand how labs are performing and interpreting TMB. It’s still unclear what’s a TMB-high vs TMB-low result? Is 10 an arbitrary cutoff point? Is TMB determined based on one clinical test, or the ratio between two TMB tests?
We’re working with International Quality Network for Pathology (IQN Path) to scope the development of an EQA for TMB testing. I don’t see it happening quickly because labs are struggling with how to measure TMB and clinicians are struggling with how to interpret TMB test data.
Q: What can we do as a community to help create the evidence of clinical validity, utility, and cost effectiveness for NGS oncology panels?
MHT: We are a global community of public and private stakeholders, and international EQA agencies that come together to achieve quality results and reduce patient costs. Investigator-initiated trials (IIT), pharma-initiated trials, or government supported studies will enable us to obtain clinical validity and utility data. However, the question of cost effectiveness is challenging. It can vary depending on different reimbursement situations in different regions. Every region usually works out its own cost-effectiveness analysis. A good analysis from the UK NICE, which is a well known authority in the field for cost-effectiveness analysis, would still be evaluated in other countries with context in mind.
LJ: It all boils down to accurate data. We need analytical validity before we can have clinical validity, and we need clinical validity before we can have clinical utility. However, we still don’t have full confidence in NGS data, which is analytical validity, to feel confident about clinical validity.
The germline variant community has the ClinVar database, which has grown rapidly and become useful for defining clinical validity. Previously, we had the Human Gene Mutation Database (HGMD), which was a manually curated database of variant calls in the literature. Unfortunately, about two-thirds of the calls in HGMD were overcalls, so the database wasn’t useful. In contrast, labs upload reports to ClinVar with data about relationships between disease-associated variants and phenotypes. It’s a free archive that’s easy to query and consists of useful, well-annotated, and well-classified variant information.
I think it is possible for the somatic variant community to create a similar database. We’re seeing it to some extent with the cBioPortal and OncoKB databases. The challenge will be to create a standard structure for importing data.
SP: For clinical validation, we need harmonization of data standards and nomenclature to make sure that we’re all talking about the same thing, ie which variant for which phenotype. None of the databases that we’ve been talking about will work otherwise.
Writing best practice guidelines is something I feel very strongly about as an organization. We are a community of laboratories trying to do the best for the patient and get the right result. We need to work together to improve guidance for laboratories in implementing NGS panels.
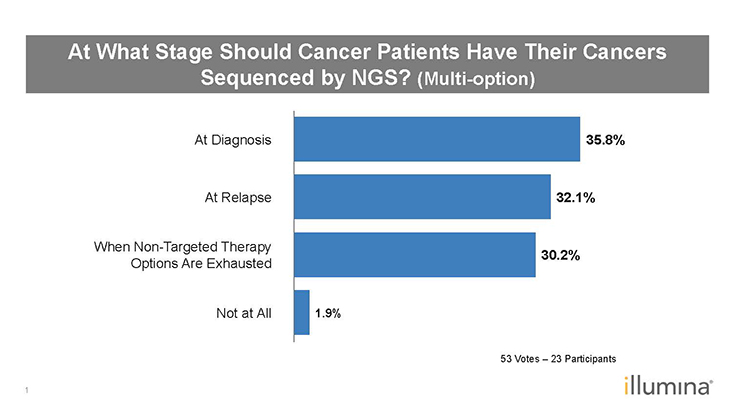
Figure 3: Poll Results–At What Stage Should Cancer Patients Have Their Cancers Sequenced by NGS?1
Q: What changes are necessary to support wide adoption of NGS outside of specialist reference centers?
MHT: One key aspect of mainstream market testing is at the primary care physician level. In the United States, Regeneron Genetics Center and the Geisinger Health System have collaborated with a goal of enrolling 250,000 people for exome sequencing focusing on well understood preventable illnesses.2 The goal is to sequence and analyze 100,000 exomes per year. In some ways, we are entering the realm of scientific fortune telling. The meaning of that information is really evolving alongside the need for clinician education. We don’t always do that well. In fact, publications have shown that within a well resourced country like the United States, two-thirds of BRCA testing is performed without genetic counseling. How do we, as a medical community, make sure that people understand and interpret the results correctly? Shouldn't the effort be focused on ensuring that the clinical and patient communities are sufficiently educated to have the right conversations on disease prevention? The Geisinger Health System is doing the right thing in pushing the envelope and creating a basis to drive the discussion with people.
SP: Genetics will become much more mainstream. Look at the growth of NGS in the last decade. The oncology sphere alone has grown dramatically in the last 5–6 years. I foresee a significant increase in point-of-care testing, with genetics performed at the patient bedside. These quick tests could inform a faster patient care decision-making process. As genetics becomes a part of mainstream medicine we need to see more clinician education, especially for the general practitioners. General practitioners (GP) are already becoming involved in the process of ordering genetic tests.
LJ: Direct-to-consumer and bedside genetic testing have the same thing in common, the need for genetic counseling to help patients and their families understand test results. For example, if you perform a germline test and identify homozygous Factor V Leiden, it might not have as much impact on the patient as it does on their sibling who is a middle-age female smoker. Are we going to perform the test, identify this variant, and fail to inform other family members that they might possess the variant and what the health implications could be? If someone receives a BRCA test, do we have an obligation to inform the rest of their family? Even with somatic variants we’re looking at germline variants and there might be incidental findings.
The challenge is that we don’t have the genetic counseling resources to advise families and patients about these scenarios. We need more education on the primary care side, among the primary care physicians, physician assistants (PA), registered nurses (RN), and advanced practice nurses (APN).
SP: The paradigm is already there because BRCA testing is the first mainstream genetic test that has a crossover between the somatic and the germline perspective. It’s a concern in terms of clinical practice. There’s so much variation in the clinical implementation of that type of test, making education really important. I see so many labs just doing a somatic mutation test and completely forgetting about the germline, and therefore the familial implications of finding a mutation.
LJ: For example, in pediatrics, about 10–15% of kids with cancer have a germline predisposition variant. That has implications for the whole family. You cannot simply perform genetic testing on a child without considering that.
Summary
NGS has and will become an essential tool in molecular diagnostic laboratories throughout the world. Clinicians and pathologists understand the potential value and advantages provided by single and multianalyte NGS cancer panel testing in diagnosing and treating patients. Analytical validity data, robust clinical variant databases, NGS panel test reimbursement strategies, and best practice guidelines to ensure clinical validity will be necessary before NGS panels will become routine clinical tests worldwide.
Interested in Reading More?
In January 2019, Illumina announced that the FDA granted prioritized review and resources for its pan-cancer assay.
Learn about Illumina's new liquid biopsy and high-throughput assays here.
References
- Attendee polling response data. Implementation of NGS in Precision Oncology meeting. Singapore. August 17, 2018.
- Regeneron. About the Regeneron Genetics Center and Geisinger Health System Collaboration. www.regeneron.com/sites/all/themes/regeneron_corporate/files/science/RGC_FactSheet_GHSCollaboration_Final.pdf. Accessed October 15, 2018.
