
COVIDSeq Assay (96 samples)
This low- to mid-throughput NGS assay enables labs of any size to identify and track the emergence and prevalence of novel SARS-CoV-2 variants.
Library prep and enrichment kit that delivers highly sensitive, comprehensive respiratory pathogen identification and antimicrobial resistance insights.
Assay time
Hands-on time
Input quantity
The Respiratory Pathogen ID/AMR Enrichment Panel (RPIP) Kit is a next-generation sequencing (NGS)-based respiratory pathogen panel and library prep and enrichment kit that targets > 280 respiratory pathogens and > 2000 antimicrobial resistance (AMR) markers in a single assay, providing both pathogen identification and associated AMR information.
Together with DRAGEN Microbial Enrichment Plus, the flexible and scalable sample-to-results workflow delivers a rapid, cost-effective solution for identifying pathogens associated with respiratory tract infections in clinical research settings.
| Assay time | < 9 hr library prep time |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Automation details | Explore available automation methods |
| Content specifications | Detects respiratory pathogen DNA and RNA simultaneously and profiles antimicrobial resistance (AMR) gene expression concurrently. |
| Description | Identify respiratory infections and co-infections, detect antimicrobial resistance markers, and perform strain typing of critical pathogens (SARS-CoV-2 and Flu A/B viruses) to study viral evolution and transmission. |
| Hands-on time | < 2 hr library prep time |
| Input quantity | Based on volume (not concentration dependent) |
| Instruments | MiSeq System, NextSeq 550 System, MiniSeq System, MiSeq i100 Plus System, MiSeq i100 System |
| Mechanism of action | On-bead tagmentation followed by a single hybridization step |
| Method | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment |
| Multiplexing | Up to 384 samples in a single run with unique dual indexes |
| Nucleic acid type | DNA, RNA |
| Species category | Fungal, Virus, Bacteria |
| Species details | Detects respiratory pathogens (180+ bacteria, 50+ fungi, and 40+ viruses, including SARS-CoV-2) and antimicrobial resistance alleles (1200+). |
| Technology | Sequencing |
In addition to the Respiratory Pathogen ID/AMR Enrichment Panel Kit, you'll need a sequencing instrument, compatible sequencing reagents, and additional products listed in the user guide in the Documentation section.
Respiratory Pathogen ID/AMR Enrichment Panel Kit
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| MiniSeq System | MiniSeq Rapid Kit: 20 samples per run based on 1M single reads per sample. MiniSeq High-Output Kit: 25 samples per run based on 1M single reads per sample. |
Rapid Kit: 1 × 100 bp. High-Output Kit: 1 × 75 bp |
| MiSeq System | MiSeq v3 Kit: 25 samples per run based on 2M paired-end reads per sample. MiSeq v2 Kit: 15 samples per run based on 2M paired-end reads per sample. |
MiSeq v3 Kit: 2 × 75 bp MiSeq v2 Kit: 2 × 150 bp |
| NextSeq 550 System | NextSeq 550 High-Output Kit: 384 (available indexes) samples per run based on 2M paired-end reads per sample. NextSeq 550 Mid-Output Kit: 130 samples per run based on 2M paired-end reads per sample. |
NextSeq 550 High-Output Kit: 2 × 75 bp. NextSeq 550 High-Output Kit: 2 × 150 bp. NextSeq 550 Mid-Output Kit: 2 × 75 bp. NextSeq 550 Mid-Output Kit: 2 × 150 bp. |
NGS can identify novel coronavirus variants, track COVID-19 transmission, and more. Compare NGS methods for various coronavirus sequencing goals.
Genomic surveillance of infectious diseases
NGS supports effective genomic surveillance strategies through identification of novel strains of coronavirus and other emerging infectious disease agents.
| Respiratory Pathogen ID/AMR Enrichment Panel Kit | Illumina Respiratory Virus Enrichment Kit | |
|---|---|---|
| Assay time | < 9 hr library prep time | < 9 hr library prep time |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) |
| Automation details | Explore available automation methods | Explore available automation methods |
| Content specifications | Detects respiratory pathogen DNA and RNA simultaneously and profiles antimicrobial resistance (AMR) gene expression concurrently. | Targets and characterizes ~40 common respiratory viruses, including SARS-CoV-2, influenza A and B viruses, adenovirus, rhinovirus, respiratory syncytial virus, and other common respiratory viruses. |
| Description | Identify respiratory infections and co-infections, detect antimicrobial resistance markers, and perform strain typing of critical pathogens (SARS-CoV-2 and Flu A/B viruses) to study viral evolution and transmission. | A streamlined workflow for detecting and analyzing SARS-CoV-2 and other common respiratory viruses. The agnostic design allows for widespread identification of pathogenic viruses across multiple sample types of interest. |
| Hands-on time | < 2 hr library prep time | < 2 hr library prep time |
| Input quantity | Based on volume (not concentration dependent) | 10-100 ng |
| Instruments | MiSeq System, NextSeq 550 System, MiniSeq System, MiSeq i100 Plus System, MiSeq i100 System | MiSeq System, NextSeq 550 System, NextSeq 2000 System, MiniSeq System, MiSeq i100 Plus System, MiSeq i100 System |
| Mechanism of action | On-bead tagmentation followed by a single hybridization step | On-bead tagmentation followed by a single hybridization step |
| Method | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment |
| Multiplexing | Up to 384 samples in a single run with unique dual indexes | Up to 384 samples in a single run with unique dual indexes |
| Nucleic acid type | DNA, RNA | DNA, RNA |
| Species category | Fungal, Virus, Bacteria | Human, Virus |
| Species details | Detects respiratory pathogens (180+ bacteria, 50+ fungi, and 40+ viruses, including SARS-CoV-2) and antimicrobial resistance alleles (1200+). | Detects common respiratory viruses, including recent influenza virus strains and SARS-CoV-2. The panel includes probes to human targets as a quality feature, and therefore detects those human targets in every sample assayed. |
| Technology | Sequencing | Sequencing |
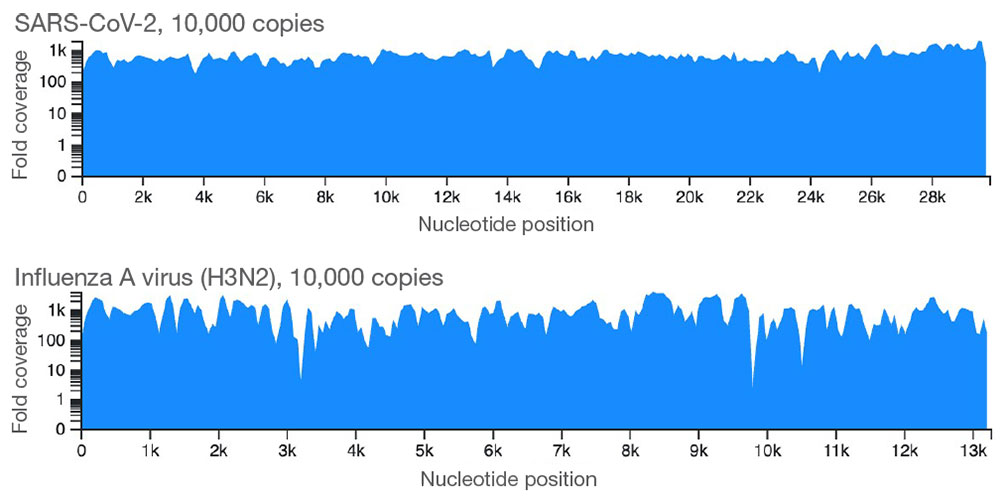

Target enrichment probes and the Respiratory Pathogen ID/AMR Panel Kit workflow provide uniform coverage of the two viruses in the panel that are tiled completely with probes.
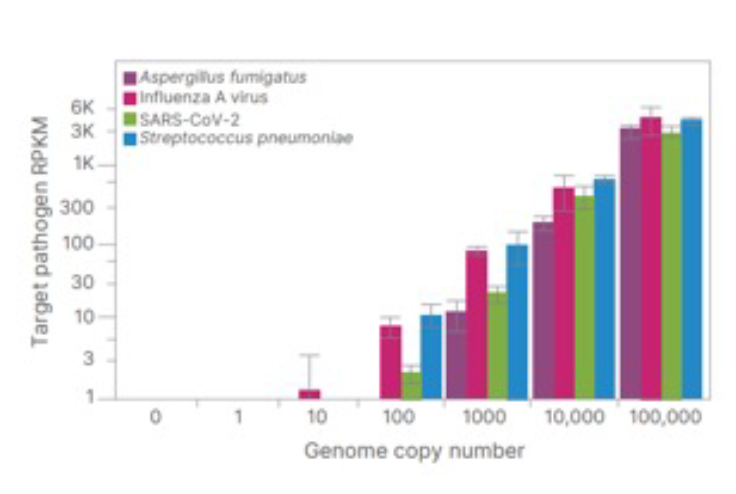

Using the Respiratory Pathogen ID/AMR Panel Kit, pathogens were detectable down to 100 genome copies except for A. fumigatus. Detection of Influenza A was more sensitive and detected to 10 genome copies.
Respiratory Pathogen ID/AMR Enrichment Kit Set A (RUO) (96 indexes, 96 samples)
20047050
Includes all reagents necessary for library prep and enrichment including Illumina RNA Prep with Enrichment kit reagents, IDT for Illumina DNA/RNA UD Indexes Set A, and Respiratory Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Respiratory Pathogen ID/AMR Enrichment Kit Set B (RUO) (96 indexes, 96 samples)
20046969
Includes all reagents necessary for library prep and enrichment including Illumina RNA Prep with Enrichment kit reagents, IDT for Illumina DNA/RNA UD Indexes Set B, and Respiratory Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Respiratory Pathogen ID/AMR Enrichment Kit Set C (RUO) (96 indexes, 96 Samples)
20047051
Includes all reagents necessary for library prep and enrichment including Illumina RNA Prep with Enrichment kit reagents, IDT for Illumina DNA/RNA UD Indexes Set C, and Respiratory Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Respiratory Pathogen ID/AMR Enrichment Kit Set D (RUO) (96 indexes, 96 Samples)
20047052
Includes all reagents necessary for library prep and enrichment including Illumina RNA Prep with Enrichment kit reagents, IDT for Illumina DNA/RNA UD Indexes Set D, and Respiratory Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
We generally recommend starting with 1M single reads, or 2M paired-end reads per sample using a minimum of 1 × 100 bp or 2 × 75 bp sequencing chemistries. These numbers are a starting point and should be optimized based on sample types and target abundance.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.