
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
This panel supports characterization and genomic surveillance of 225 known and novel coronaviruses from humans and animal reservoirs.
Assay time
Hands-on time
Input quantity
The Pan-Coronavirus Panel (Pan-CoV) allows for detection and sequencing of known human and animal coronaviruses and closely related novel coronaviruses with the potential to become infectious to humans. It includes hybrid–capture oligos that span across reference genomes of multiple coronavirus strains and various known animal hosts.
When combined with Illumina RNA Prep with Enrichment, the Pan-Coronavirus Panel enables whole-genome sequencing (WGS) of 225 known human and animal coronaviruses. The targeted enrichment approach captures mutations and variants throughout the viral genome, enabling sequencing of novel coronavirus strains and detection of recombination between strains.
The panel can be used for multiple applications, including:
The Pan-Coronavirus Panel can integrate into a comprehensive workflow that includes library preparation, target enrichment, next-generation sequencing (NGS), and powerful data analysis, providing a simple means to obtain robust and reliable viral results.
| Assay time | < 9 hr library prep time |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Description | The Pan-Coronavirus Panel is part of an integrated workflow that allows for the detection and whole-genome sequencing of over 200 known and novel coronavirus strains in various animal hosts. |
| Hands-on time | < 2 hr library prep time |
| Input quantity | 10-100 ng |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System |
| Mechanism of action | On-bead tagmentation followed by a single hybridization step |
| Method | Whole-genome sequencing, Target enrichment |
| Multiplexing | Up to 384 samples in a single run with unique dual indexes |
| Nucleic acid type | RNA |
| Species category | Virus |
| Strand specificity | Non-stranded |
| Technology | Sequencing |
Illumina RNA Prep with Enrichment, (L) Tagmentation (16 or 96 Samples) needs to be ordered separately as well as index adapters. Illumina DNA/RNA Indexes Set A, B, C, or D can be used.
We recommend ordering the Illumina DNA/RNA UD Indexes Sets A-D (20091654, 20091656, 20091658, 20091660) since the IDT for Illumina DNA/RNA UD Indexes Sets A-D (20027213, 20027214,20042666, 20042667) are being discontinued.
Target enrichment through hybrid–capture allows for detection and sequencing of 225 known human and animal coronaviruses and closely related novel coronaviruses for surveillance of animal reservoirs, tracking transmissions, and therapeutic and vaccine development.
Pan-Coronavirus Panel
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| MiSeq System | 6–12 samples using MiSeq Reagent Kit v2 (1–2M reads/sample) |
2 × 150 bp |
| NextSeq 550 System | 65–130 samples using NextSeq 500/550 Mid-Output Kit (1–2M reads/sample) |
2 × 150 bp |
| NextSeq 1000 System | 100–200 samples using P2 Reagent Kit (1–2M reads/sample) |
2 × 150 bp |
| NextSeq 2000 System | 100–200 samples using P2 Reagent Kit (1–2M reads/sample) |
2 × 150 bp |
Genomic surveillance of infectious diseases
NGS supports effective genomic surveillance strategies through identification of novel strains of coronavirus and other emerging infectious disease agents.
NGS can identify novel coronavirus variants, track COVID-19 transmission, and more. Compare NGS methods for various coronavirus sequencing goals.
Use NGS to discover novel microbes, monitor outbreaks, analyze food sources, and more.
| Pan-Coronavirus Panel | Viral Surveillance Panel | Illumina Respiratory Virus Enrichment Kit | |
|---|---|---|---|
| Assay time | < 9 hr library prep time | < 9 hr library prep time | < 9 hr library prep time |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) | Liquid handling robot(s) |
| Description | The Pan-Coronavirus Panel is part of an integrated workflow that allows for the detection and whole-genome sequencing of over 200 known and novel coronavirus strains in various animal hosts. | The Viral Surveillance Panel v2 Kit provides an optimized workflow that supports viral outbreak detection and monitoring of more than 200 RNA and DNA viral pathogens. | A streamlined workflow for detecting and analyzing SARS-CoV-2 and other common respiratory viruses. The agnostic design allows for widespread identification of pathogenic viruses across multiple sample types of interest. |
| Hands-on time | < 2 hr library prep time | < 2 hr library prep time | < 2 hr library prep time |
| Input quantity | 10-100 ng | 10-100 ng | 10-100 ng |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System, MiSeq i100 Plus System, MiSeq i100 System | MiSeq System, NextSeq 550 System, NextSeq 2000 System, MiniSeq System, MiSeq i100 Plus System, MiSeq i100 System |
| Mechanism of action | On-bead tagmentation followed by a single hybridization step | On-bead tagmentation followed by a single hybridization step | On-bead tagmentation followed by a single hybridization step |
| Method | Whole-genome sequencing, Target enrichment | Targeted DNA sequencing, Whole-genome sequencing, Targeted RNA sequencing, Target enrichment | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment |
| Multiplexing | Up to 384 samples in a single run with unique dual indexes | Up to 384 samples in a single run with unique dual indexes | Up to 384 samples in a single run with unique dual indexes |
| Nucleic acid type | RNA | DNA, RNA | DNA, RNA |
| Species category | Virus | Virus | Human, Virus |
| Strand specificity | Non-stranded | Non-stranded | |
| Technology | Sequencing | Sequencing | Sequencing |
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
Sequencing Coverage Calculator
Calculator to help determine the reagents and sequencing runs needed to arrive at desired coverage for your experiment.


Coronavirus strains covered in panel
Pan-Coronavirus Panel includes hybrid–capture oligos that capture 225 known human and animal coronaviruses.


Host-associated coronavirus genomes
Pan-Coronavirus Panel includes various known animal hosts.


Enrichment of Coronaviridae reads
The Pan-Coronavirus Panel successfully enriched Coronaviridae reads in all libraries tested across a range of viral copy number input.
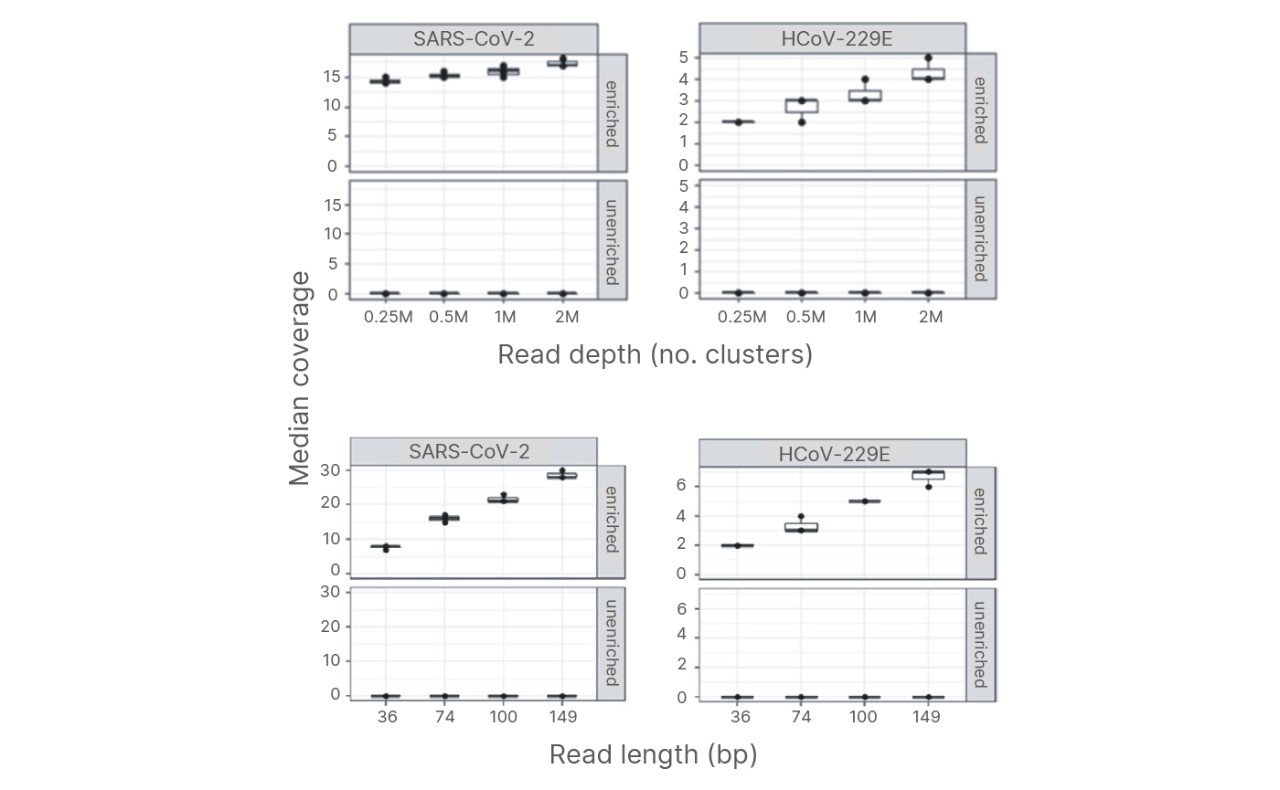

Improved coronavirus genome coverage
Median coverage of synthetic viral RNA controls (SARS-CoV-2 and HCoV229E) in 10 ng UHR after enrichment with the Pan-Coronavirus Panel. Increased read depth (top) and read length (bottom) resulted in improved genome coverage. Read lengths were trimmed to 36, 74, 100, and 149 bp.
Pan-Coronavirus Panel, RUO 96 Rxns
20088155
Includes Viral Surveillance Panel sufficient for 32, 3-plex enrichment reactions when paired with Illumina RNA Prep with Enrichment kit. Purchase library prep and enrichment kit and index adapters separately.
List Price:
Discounts:
Illumina® RNA Prep with Enrichment, (L) Tagmentation (16 Samples)
20040536
Includes reagents for preparing and enriching 16 libraries (16, 1-plex enrichment reactions). Purchase enrichment probe panel, Purification Beads and index adapters separately.
List Price:
Discounts:
Illumina® RNA Prep with Enrichment, (L) Tagmentation (96 Samples)
20040537
Includes reagents for preparing and enriching 96 libraries (32, 3-plex enrichment reactions). Purchase enrichment probe panel, Purification Beads and index adapters separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set A, Tagmentation (96 Indexes, 96 Samples)
20091654
Illumina® DNA/RNA UD Indexes Set A, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set B, Tagmentation (96 Indexes, 96 Samples)
20091656
Illumina® DNA/RNA UD Indexes Set B, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set C, Tagmentation (96 Indexes, 96 Samples)
20091658
Illumina® DNA/RNA UD Indexes Set C, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set D, Tagmentation (96 Indexes, 96 Samples)
20091660
Illumina® DNA/RNA UD Indexes Set D, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
The panel is built with enrichment probes that allows for most variants to be captured effectively. In addition, the Pan-Coronavirus Panel was designed to enrich for 225 coronavirus genotypes from various animal sources and expected to capture most novel coronaviruses. For example, the Pan-Coronavirus Panel was designed before the Omicron variant but was still shown to enrich for the entire genome with high coverage (see data sheet for details).
Yes, the Pan-Coronavirus Panel was designed to work on environmental samples and samples from various animal sources. In fact, the panel is to be used for sequencing of coronviruses from diverse sample types such as bats, wastewater, and any potential coronavirus host. To maximize success with the panels, use the highest quality RNA you can obtain.
There is a free DRAGEN Microbial Enrichment Plus that can assemble contigs and explore genome coverage/depth, in addition to other outputs.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.