Ribosome Profiling
What is Ribosome Profiling?
Ribosome profiling is a method based on deep sequencing of ribosome-protected mRNA fragments. Ribosome profiling, also known as Ribo-Seq (ribosome sequencing) or ART-Seq (active mRNA translation sequencing), provides a “snapshot” of all the ribosomes active in a cell at a specific time point. This information can help researchers determine which proteins are being actively translated in a cell.
Ribosome profiling analysis enables systematic monitoring of cellular translation processes and prediction of protein abundance. Determining what regions of a message are being translated can help define the proteome of complex organisms. In combination with massively parallel high-throughput next-generation sequencing (NGS), ribosome profiling with Ribo-Seq allows detailed and accurate in vivo analysis of protein production.

Advantages of Ribosome Profiling
By identifying transcripts undergoing active translation, Ribo-Seq can provide visibility into an important aspect of gene regulation.
- Investigate translational control and measure gene expression
- Identify translation start sites
- Determine the rate of protein synthesis
- Predict protein abundance
Generating Sequencing Libraries for Ribosome Profiling
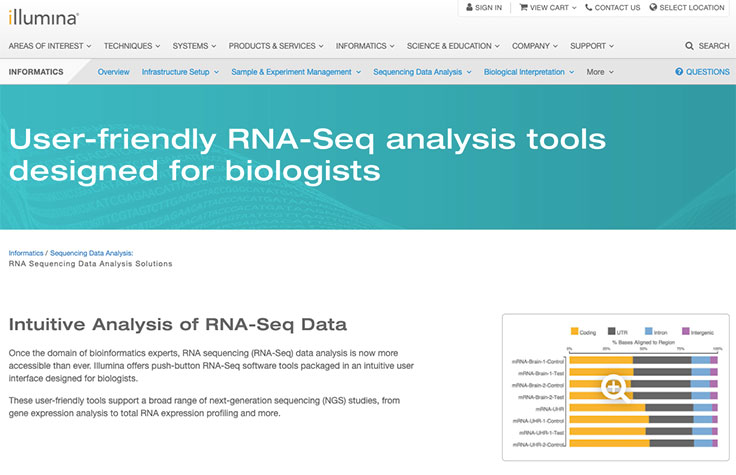
Ribosome profiling with Ribo-Seq uses size-exclusion chromatography. This method streamlines the steps to convert ribosome-protected RNA fragments into libraries compatible with sequencing on Illumina instruments. It is a simple, rapid, and scalable method that does not require special equipment. In contrast, other methods to isolate polysomes and monosomes rely on several hours of ultracentrifugation using either a sucrose gradient or a cushion.
Featured Products
NextSeq 2000 System

Groundbreaking benchtop sequencers allow you to explore new discoveries across a variety of current and emerging applications, with higher efficiency and fewer restraints.
View SystemBaseSpace Sequence Hub
Simplify your bioinformatics with an economical and powerful cloud computing environment to manage, analyze, and share data.
Learn MoreRibosome Profiling Protocol
The TruSeq Ribo Profile (formerly known as ART-Seq) kits, which have been discontinued, are based on the following published protocol for ribosome profiling. This publication provides detailed information about the reagents required for the assay and where they can be purchased, as a possible alternative to TruSeq Ribo Profile.
Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8);1534-1550.
Read Publication
Related Solutions
Cancer Research: Gene Deregulation
Ribosome sequencing can provide critical insight into gene deregulation in cancer. This method, as well as gene expression analysis, can help researchers gain insight into functional changes occurring during cancer development. Learn more about cancer gene expression studies.
Complex Disease Research: Translational Regulation
Ribosome sequencing further bridges the gap between genomics/transcriptomics and proteomics, providing visibility to an important layer of post-transcriptional regulation and potential new insights into the mechanisms driving complex diseases. Learn more about complex disease genomics.
TruSeq Ribo Profile kits have been discontinued
Interested in receiving newsletters, case studies, and information on sequencing methods?
Enter your email address.
Additional Resources
*Data calculations on file. Illumina, Inc., 2015