Next-generation Sequencing Glossary
NGS glossary
Guide to NGS terminology
Use our next-generation sequencing glossary to view definitions of key terms and clarify important concepts as you plan your sequencing project.
The critical difference between Sanger sequencing and NGS is sequencing volume. While the Sanger method only sequences a single DNA fragment at a time, NGS is massively parallel, that is, sequencing millions of fragments simultaneously per run. This process translates into sequencing hundreds to thousands of genes at one time. NGS also offers greater discovery power to detect novel or rare variants with deep sequencing.
Glossary of common NGS terms
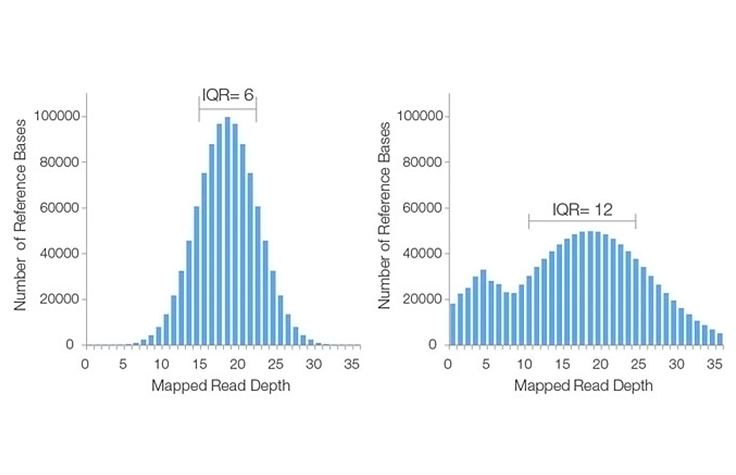
The average number of sequenced bases that align to, or “cover,” known reference bases. For example, a whole genome sequenced at 30× coverage means that, on average, each base in the genome was sequenced 30 times. At higher levels of coverage, base calls can be made with a higher degree of confidence.
Learn More
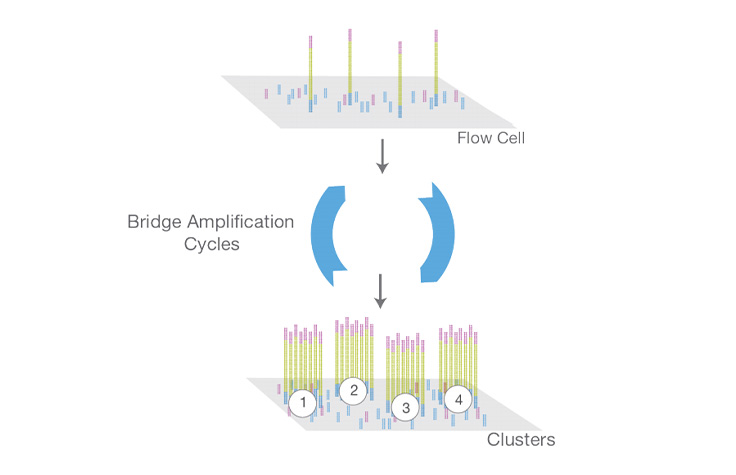
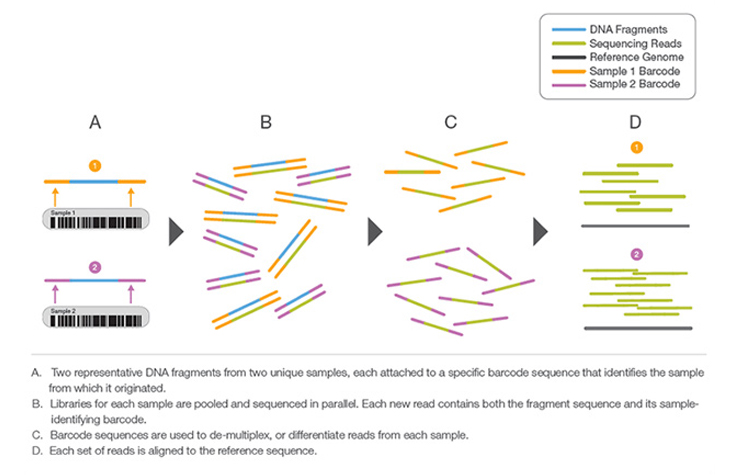
Library Multiplexing Overview

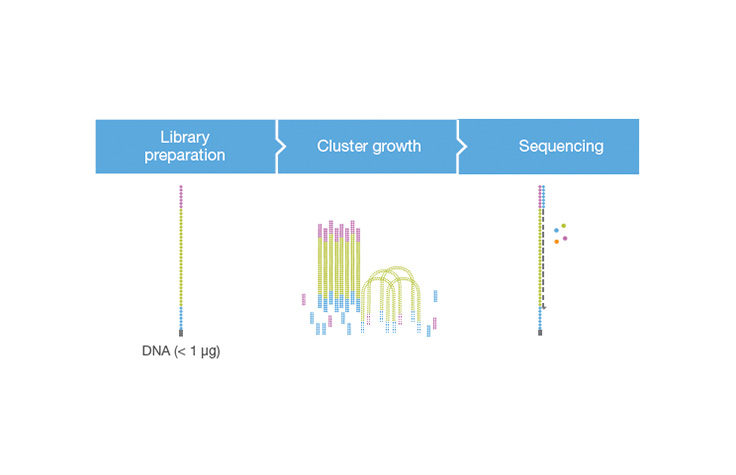
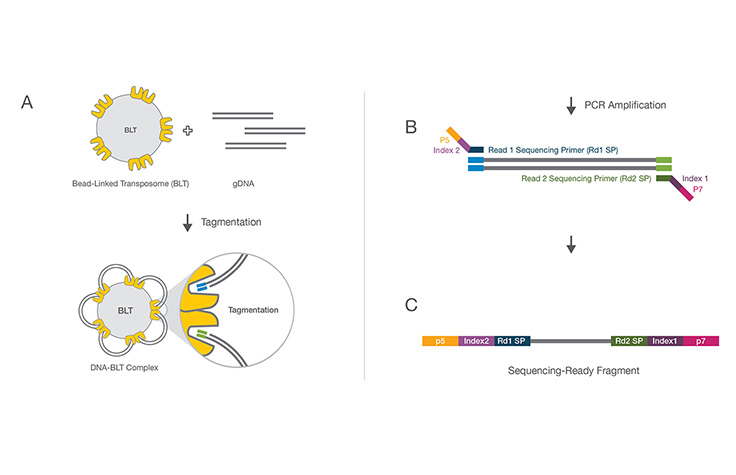
A molecular biology protocol that converts a genomic DNA sample (or cDNA sample) into a sequencing library, which can then be sequenced on an NGS instrument. The first step in library preparation is random fragmentation of the DNA sample, followed by ligation of 5ʹ and 3′ adapters to each DNA fragment. Alternatively, “tagmentation” combines the fragmentation and ligation reactions into a single step and greatly increases the efficiency of the library preparation process.
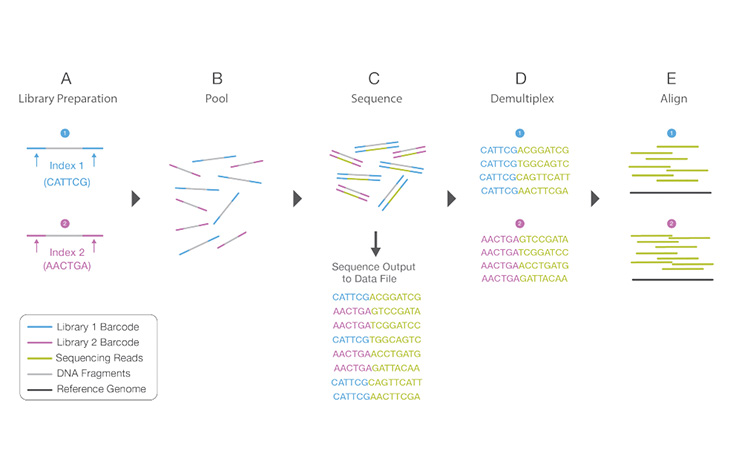
Learn MoreA process by which unique short DNA sequences, or “indexes,” are added to each DNA fragment during library preparation. The unique sequences allow many libraries to be pooled together and sequenced simultaneously. Sequencing reads from pooled libraries are identified and sorted computationally before final data analysis. Library multiplexing is a useful technique when working with small genomes or targeting genomic regions of interest. Multiplexing can exponentially increase the number of samples analyzed in a single run, without drastically increasing run cost or run time.
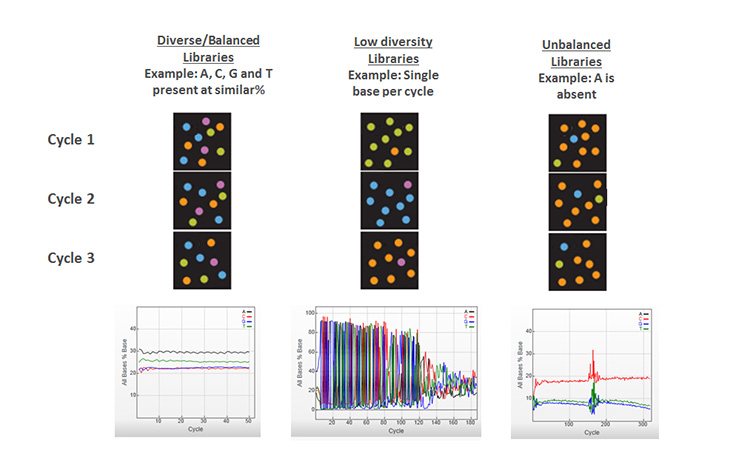
Learn MoreHaving equal proportions of A, C, G, and T nucleotides at each base position across all the DNA fragments in a sequencing library. Color balance is required for effective image analysis on Illumina sequencing systems. Therefore, most Illumina library preparation workflows include a random fragmentation step, which generates the necessary sequence diversity at each base position in the library.
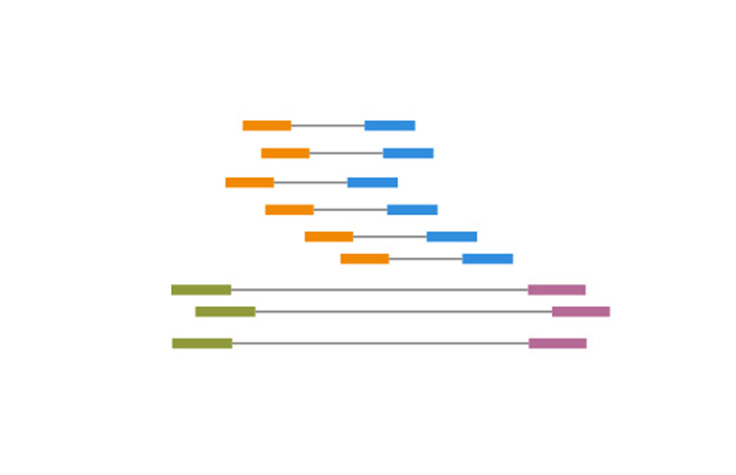
Illumina KnowledgeA process of sequencing from both ends of a DNA fragment in the same run.
Learn More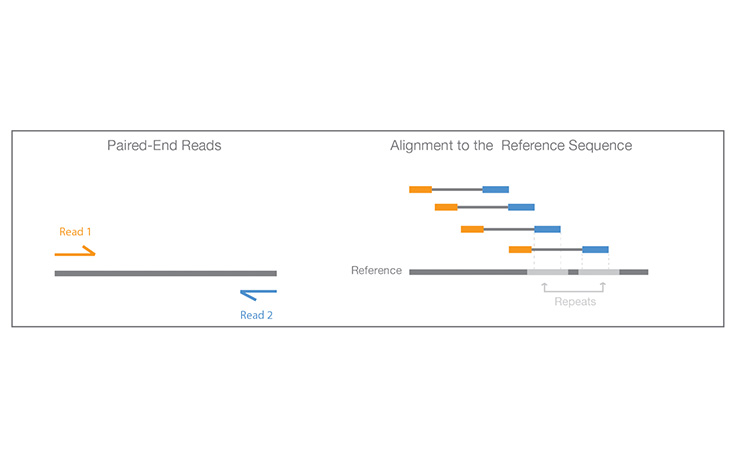
Paired-End Sequencing and Alignment

A metric in NGS that predicts or estimates the probability of an error in base calling. A quality score (Q-score) serves as a compact way to communicate very small error probabilities. A high Q-score implies that a base call is more reliable and less likely to be incorrect.
Learn More1 megabase = 1,000,000 bases.
1 gigabase = 1,000,000,000 bases.
Pairs of indexes such that every i5 index and every i7 index are used only one time. With unique dual indexes, you can identify and filter indexed hopped reads, which provides higher confidence in multiplexed samples.
Read BulletinA widely used sequencing method that targets only the protein-coding region of the genome (the exome).
Learn MoreA comprehensive method that provides a base-by-base view of the entire genome.
Learn More
Getting Started with NGS eBook
Curious about using NGS for your research? While adopting a new technology may seem intimidating, we’ve created a comprehensive, yet easy-to-follow guide for bringing NGS into your lab. You’ll learn about NGS methods, workflows, data analysis solutions, additional glossary terms, and find a step-by-step guide to getting started with NGS.
Download eBookDownload the New to NGS eBook
Additional resources
NGS for beginners
Want to learn more about NGS? This page offers simple, clear explanations of next-generation sequencing and its numerous benefits over conventional methods.
NGS tutorials
Browse our comprehensive list of tutorials designed to help you understand key concepts in NGS. We’ll guide you through best practices for library prep, sequencing, and data analysis.
Online training
Access on-demand training for instruments, library prep kits, software, and more.