Emedgene brochure
Understand how automating insights can help you confidently scale your data operations.
Unlock genomic insights for hereditary disease
Emedgene variant interpretation software streamlines your tertiary analysis workflows for rare disease genomics and other germline research applications
Your email address is never shared with third parties.
Emedgene software is designed to accelerate the time and certainty in user-defined variant interpretation, prioritization, curation, and research report generation from sequencing and microarrays.
Enable greater efficiency from your tertiary analysis workflows with explainable AI (XAI) and automation supporting genomes, exomes, virtual panels, targeted panels, and microarrays. From dozens of samples to populations, save valuable time per subject and per project.
Unify your wet lab and dry lab to simplify and secure your complete sequencing or microarray workflow. Integrate with DRAGEN secondary analysis for accurate, comprehensive, and efficient variant calling. Speed user-driven, variant interpretation by up to 75% per subject from single sample workflows to population scale studies.
Confidently keep pace with evolving science, technology, and test volumes with up-to-date knowledge sources, automated curation capabilities, and a team of experts to support your journey.
Increase throughput without increasing headcount using explainable AI (XAI) and automated workflows. Speed user-driven, variant interpretation by up to 75% per subject from single sample workflows to population scale studies.
Broaden your analysis to whole-genome sequencing (WGS) or whole-exome sequencing (WES), or standardize virtual panels on a backbone assay. Analyze various variant types—SNVs, indels, short tandem repeats (STRs), copy number variants (CNVs), other structural variants, and mtDNA. Unify your variant insights from cytogenetic microarrays on the same platform.
Whether you are conducting rare disease or other genetic disease research, hereditary risk assessment, carrier screening, healthy population screening, pharmacogenomics, cytogenetics, or more, implement a high-throughput WGS, WES, virtual panel, targeted panel, or microarray workflow that is integrated into your lab's digital ecosystem.
Leverage the power of collaboration to share knowledge across a private network of labs that you define and control.
Meng L, Attali R, Talmy T, Regev Y, Mizrahi N, Smirin-Yosef P, Vossaert L, Taborda C, Santana M, Machol I, Xiao R, Dai H, Eng C, Xia F, Tzur S. Evaluation of an automated genome interpretation model for rare disease routinely used in a clinical genetic laboratory. Genet Med. 2023 Jun. PMID: 36939041.
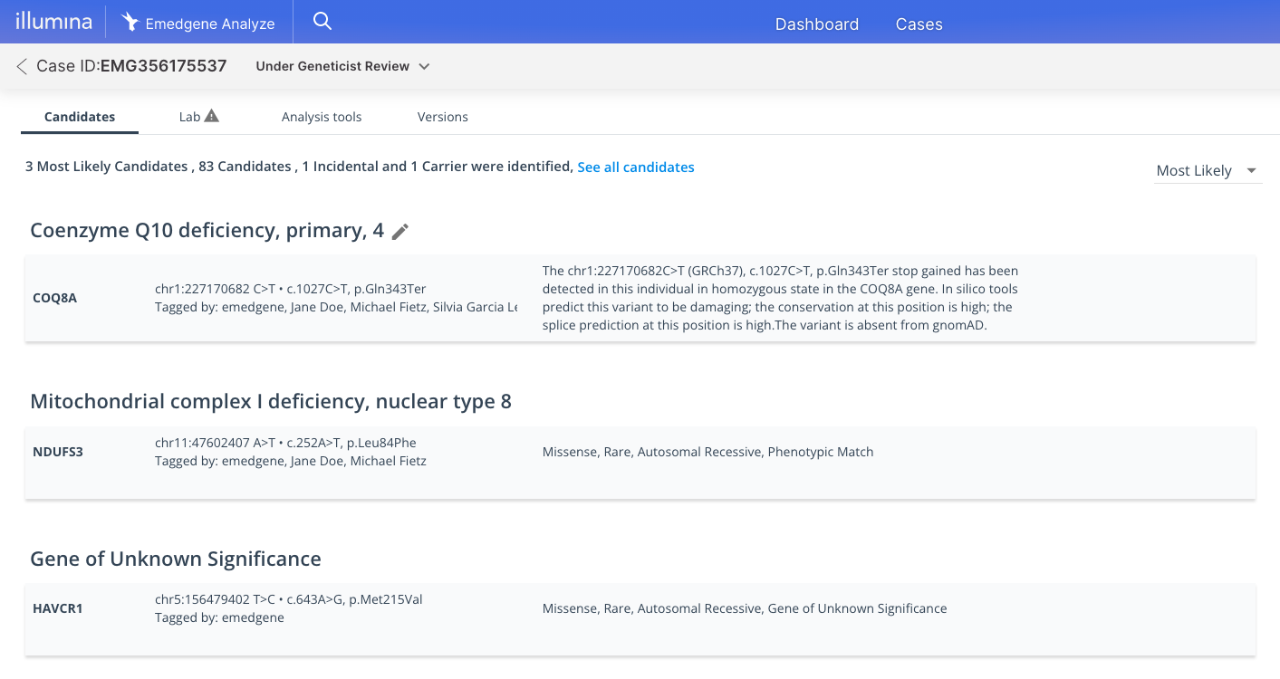

With 97% accuracy1 in prioritizing relevant insights, AI can suggest variants in complex data sets that typically require hours of manual review.
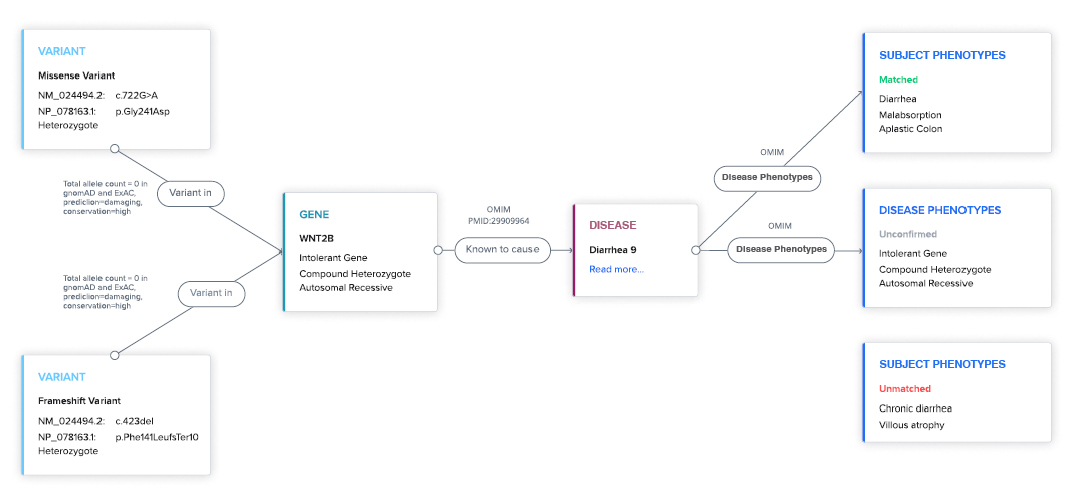

Transparent logic. Every AI hypothesis is backed by literature and database sources.
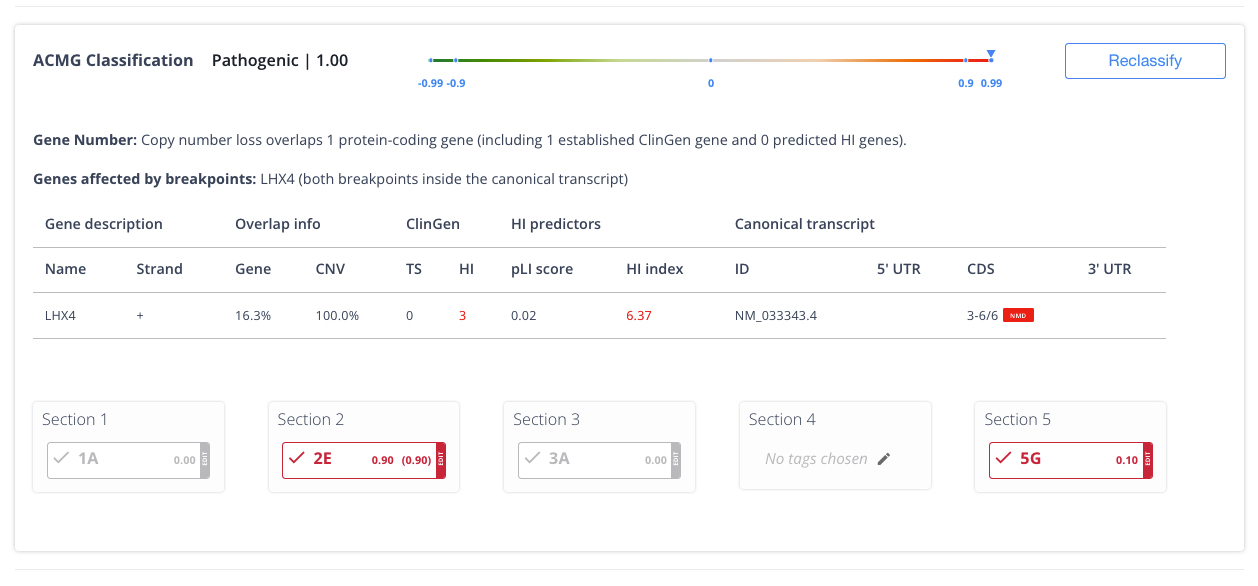

Time-saving automated ACMG classifications for SNV, indel, CNV, and SV deletions/duplication variants.
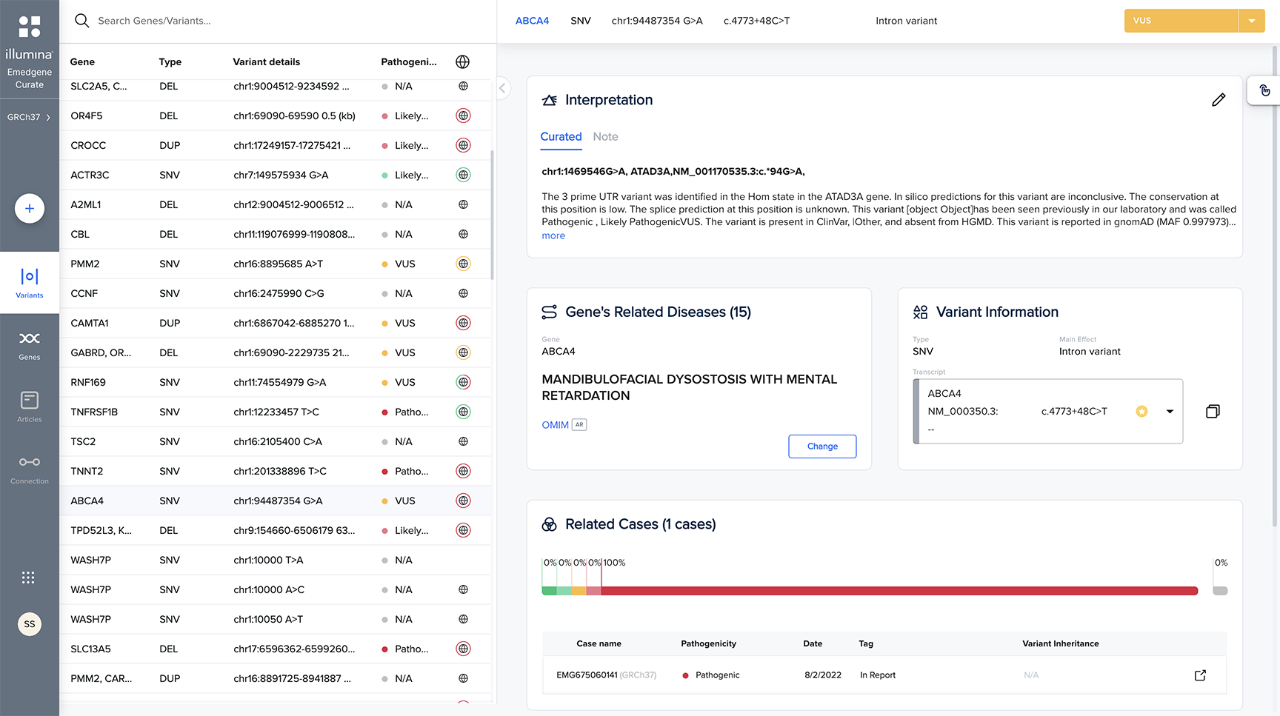

Maximize use and reuse of your organization’s curated knowledge. Share across a private network of connected labs.
Understand how automating insights can help you confidently scale your data operations.
Overview of the automated insights solution with AI prioritization that can streamline dry lab workflows for WGS, WES, virtual panels, and targeted panels.
Learn how Emedgene employs key security and privacy features, coupled with compliance certifications, to protect sensitive NGS data.
Emedgene’s machine learning simplifies the highly complex task of variant analysis, allowing us to handle more tests every day.
Dr. Linyan Meng from Baylor College of Medicine presents the results of a research study demonstrating the utility of machine learning for interpretation in a 180-sample cohort. By automating variant prioritization and classification processes, machine learning technologies support eliminating the genomic data interpretation bottleneck.
WGS is the most comprehensive method for genetic disease testing and is increasingly applied to rare disease and other hereditary disease research.
Virtual panels or “slices” can be created from a more comprehensive “backbone” assay that is standardized in the lab, such as WGS or WES.
WES evaluates the exons, or coding regions of the genome. WES can also serve as a standardized backbone assay for virtrual gene panels.
Analyze specific genes that are important for a hereditary disease or condition.
NGS technology is helping to drive breakthroughs in genetic disease testing by early detection and diagnosis.
Artificial intelligence models that can explicitly demonstrate their logic and expose potential biases, known as explainable AI, are necessary.
Hear a customer use case of explainable AI-powered variant interpretation and automation to hasten and scale their operations.
As an alternative or complement to sequencing, assess structural variation with microarrays and analyze using Emedgene.

Maximize genomic insights with DRAGEN secondary analysis, learn about the latest updates, read FAQs, and find product support.

Operationalize informatics and drive scientific insights with Illumina Connected Analytics. Contact us for information on pricing, subscriptions, and more.
Understand what a LIMS is and how it can help labs with sample tracking, workflow automation, and downstream data analysis.
References
Discuss your workflow to learn how you can streamline your NGS operations and power your lab for growth using Emedgene.
Your email address is never shared with third parties.