Coronavirus Software

Robust and Efficient Coronavirus Software Tools
To support scientists with coronavirus-related genomic data analysis and sharing, Illumina has developed a suite of tools to help identify mutations, characterize novel strains, examine the virus, and study host responses.
These tools allow researchers to identify the SARS-CoV-2 viral sequence in their samples, detect mutations, examine host immune responses, and contribute their findings to critical public databases.
See how our software tools help:
Illumina SARS-Cov-2 NGS Data Toolkit
| Novel Pathogen Detection |  |
DRAGEN Metagenomics App | ||
|---|---|---|---|---|
| Detection and Surveillance |  |
DRAGEN RNA Pathogen Detection App |  |
DRAGEN COVID Lineage App |
| Sharing and Collaboration |  |
SRA Import App |  |
GISAID Submission App |
| The Illumina SARS-CoV-2 NGS Data Toolkit is available on BaseSpace Sequence Hub. | ||||
About the Illumina SARS-CoV-2 NGS Data Toolkit
The suite of coronavirus software analysis solutions for detection and surveillance of SARS-CoV-2 is free of charge on BaseSpace Sequence Hub. The toolkit consists of several tools built on Illumina DRAGEN Secondary Analysis, and data submission apps on BaseSpace Sequence Hub, which empower researchers to seamlessly submit their findings to public databases.
DRAGEN Coronavirus Software Tools
The Illumina SARS-CoV-2 NGS Data Toolkit leverages the speed and accuracy of DRAGEN to accelerate infectious disease surveillance and outbreak response. The toolkit includes a DRAGEN Metagenomics Pipeline for identifying novel infectious disease pathogens, a DRAGEN RNA Pathogen pipeline for detection of viral pathogens, and the DRAGEN Lineage app for detection of SARS-CoV-2 mutations for epidemiology. The DRAGEN coronavirus tools can be accessed on BaseSpace Sequence Hub. Additional information about coronavirus detection and surveillance with the DRAGEN Platform can be found in this software blog post.
DRAGEN RNA Pathogen Detection Pipeline for Detection and Surveillance of Viral Pathogens
An updated version of the DRAGEN RNA Pipeline is now available to accelerate detection of viral pathogens. This pipeline helps detect SARS-CoV-2 in any DRAGEN RNA-Seq Pipeline run, regardless of application. This pipeline includes:
- Comprehensive detection, leveraging both coverage and k-mer based approaches to detect viral pathogens
- Compatibility with the Illumina respiratory virus target enrichment workflow
- Human and viral references using a combination of the human genome reference (hg38) with selected virus reference sequences
- Integrated FASTA generation for upload to public databases, such as GISAID
DRAGEN Metagenomics Pipeline for Detection and Characterization of Novel Pathogens
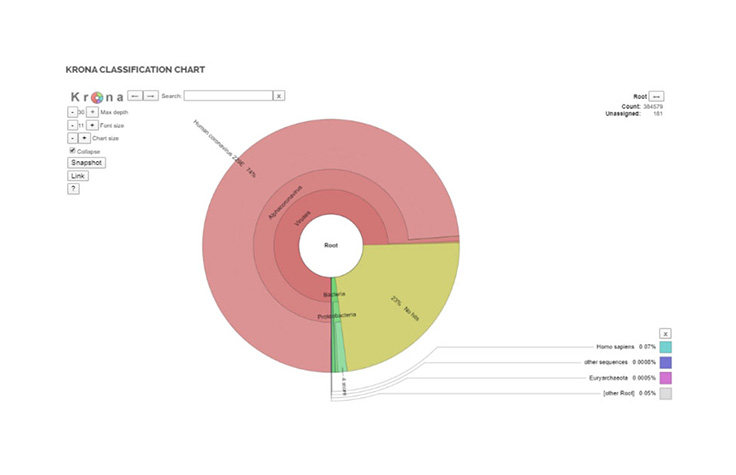
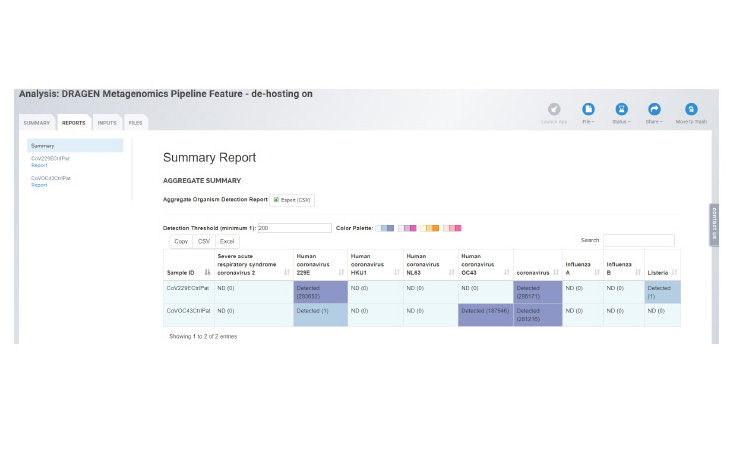
This new pipeline helps drive rapid and simplified species-level detection in shotgun metagenomics samples. The pipeline is a metagenomics classification workflow able to detect and quantify SARS-CoV-2 sequences at high sensitivity and specificity while simultaneously providing readouts for other common viral and microbial pathogens.
The pipeline leverages an updated database which includes reads specific to coronavirus and delivers a breadth of outputs including Krona plots, organism detection, and QC reports. With the ability to analyze multiple samples at once, the BaseSpace app allows researchers to look at clusters.
Access the DRAGEN Metagenomics Pipeline
View sample data generated by this pipeline:
Krona Classification ChartOrganism Detection Chart
DRAGEN COVID Lineage App
This app aligns reads to a SARS-CoV-2 reference genome and reports coverage of targeted regions. It performs:
- K-mer based detection
- Map/align, variant calling
- Consensus sequence generation
- Lineage/clade analysis using Pangolin and NextClade
Coronavirus Software for Clinical Applications
For users looking to generate clinical reporting of SARS-CoV-2 detection to diagnose COVID-19 in symptomatic patients, the DRAGEN COVIDSeq Test Pipeline is used in conjunction with the Illumina COVIDSeq Test.
For users looking to generate reports for shotgun metagenomics, the Explify platform, developed by IDbyDNA, delivers simple, powerful bioinformatics software to turn the data generated by Illumina sequencers into actionable insights. The proven, automated platform supports detection of 35 respiratory viruses, including genomic characterization of SARS-CoV-2. Contact us to learn more.
Correlation Engine for Host Response Studies
Illumina offers a collaborative environment to the COVID-19 research community to better understand critical factors that regulate the host response and help identify biomarkers for potential treatments.
How Correlation Engine can advance your research:
- Putting it into context: Correlation Engine mines thousands of genomic studies, allowing researchers to integrate their data with the world’s genomic knowledge base to generate data-driven answers.
- Accelerating research: Reduce time to discovery by leveraging Correlation Engine to query thousands of studies instantaneously and help validate hypotheses by making data-driven queries in real time.
- Staying current: Illumina has a dedicated team committed to updating Correlation Engine with curated COVID-19 data, and welcomes members of the research community to contribute their findings to help advance COVID-19 research.
- Integration: Correlation Engine is an integrated extension of our full stack of transcriptomics products to help researchers seamlessly navigate from sample to answer.
- Collaboration: Correlation Engine allows researchers to contribute and share their data with the research community to advance our understanding of the COVID-19 host response.
Learn more about Correlation Engine
Start a free Correlation Engine trial
Scientists can process samples using Clarity LIMS. Once sequencing is complete, researchers can automatically stream their data into BaseSpace Sequence Hub using the toolkit.
Powering End-to-End Solutions for
SARS-CoV-2 Detection
Illumina is making it simpler to detect and identify SARS-CoV-2 using NGS with comprehensive, integrated workflows.
Compatible with the Illumina respiratory virus target enrichment workflow, the coronavirus software toolkit empowers researchers to securely stream data directly off of our full portfolio of sequencers into BaseSpace Sequence Hub for rapid, comprehensive analysis using the DRAGEN RNA Pathogen Detection App. Once analysis is complete, researchers can submit their data to the Global Initiative on Sharing All Influenza Data (GISAID) directly from BaseSpace Sequence Hub.
For scientists looking to enhance their lab efficiency, Clarity LIMS can be used to track and monitor COVID-19 samples, delivering a comprehensive end-to-end solution.

Clarity LIMS Respiratory Virus Panel Workflow
Comprehensive Workflow Management and Sample Tracking
Clarity LIMS offers efficient sample tracking and workflow automation. The following coronavirus-related workflows are available:
- CDC COVID-19 RT-PCR workflow based on CDC recommendations
- Respiratory Virus Panel workflow
These workflows can help ensure samples are viable for analysis, allowing labs to quickly process coronavirus samples, from sample accessioning and library prep to performing QC on sequencing runs.
Learn more about Clarity LIMSCoronavirus Detection and Characterization
Explore NGS methods and find solutions to detect and characterize SARS-CoV-2 and/or other respiratory pathogens, track transmission, study co-infection, and investigate viral evolution.
Learn about coronavirus sequencing
Easy and Secure Sharing, Collaboration, and Submission to Public Databases
BaseSpace Sequence Hub is designed to facilitate secure, audit-controlled collaboration among research groups. Data management and sharing capabilities, such as workgroups, allow you to share projects and runs securely with collaborators without requiring file downloads.
The Illumina SARS-CoV-2 NGS Data Toolkit features several BaseSpace Sequence Hub apps for collaboration. The GISAID Submission App accepts a collection of VCF or FASTA files, along with a metadata CSV for the same samples. The app performs data validation before submitting data to GISAID for further QA and processing.
Download BaseSpace security and privacy infoHow to Access the Illumina SARS-CoV-2 NGS Data Toolkit
The full Illumina SARS-CoV-2 NGS Data Toolkit is available on BaseSpace Sequence Hub. The updated DRAGEN COVID-19 tools are also available on the DRAGEN Server and are being made available to developers via the DRAGEN API. To request access to the DRAGEN API, fill out the form below.
The Illumina SARS-CoV-2 NGS Data Toolkit on BaseSpace Sequence Hub
Researchers can start using the Illumina SARS-CoV-2 NGS Data Toolkit today on BaseSpace Sequence Hub, free of charge. Researchers can stream data directly from their instruments into BaseSpace’s secure cloud environment for push-button usage of the entire toolkit.
Learn more about BaseSpace Sequence HubFeatured Products
BaseSpace Sequence Hub and iCredits
BaseSpace Sequence Hub allows you to manage and analyze your data easily with a curated set of analysis apps. iCredits are the currency used to purchase Illumina genomic data storage and analysis options.

DRAGEN Bio-IT Platform
The Illumina DRAGEN Bio-IT Platform provides accurate, ultra-rapid secondary analysis of sequencing data.

NextSeq 2000 System and Reagents
The NextSeq 2000 System supports emerging and mid-throughput sequencing applications as well as a broad range of methods such as exome sequencing, target enrichment, single-cell profiling, transcriptome sequencing, and more.
To request access to the DRAGEN API, please complete the form below.
COVID-19 Host Risk and Response
Understanding host genetic differences and individual responses to the SARS-CoV-2 virus increases understanding of disease susceptibiliity and severity. Read more about the methods for host risk & immune response studies.
Learn about host genetics and immune response profiling

Additional Coronavirus Software Tools
Target Enrichment Pipeline
The DRAGEN Enrichment Pipeline can also be leveraged for detection and identification of SARS-CoV-2. The pipeline is available on BaseSpace Sequence Hub for a low per-sample cost, and can be accessed on the DRAGEN Server.
Learn more about DRAGEN Enrichment