
October 19, 2023
Before SARS-CoV-2, Aegis Sciences Corporation was a small, well-established lab with a specific niche. Founded in 1990, the Nashville-based company focused on anti-doping testing and medication compliance. Later, they started working with small and midsize pharmaceutical companies, conducting genetic testing for cancer clinical trials. Until 2020, Aegis had no infectious disease business at all.
COVID-19 changed that. When the pandemic hit, Aegis scaled up fast. The company went from six full-time lab staff to 550 in 18 months. Its COVID diagnostic testing capacity expanded from 3000 PCR tests a day at the beginning of the pandemic to 130,000 a day during the Delta wave. With this capability, Aegis was perfectly situated to help the Centers for Disease Control and Prevention (CDC) monitor changes in SARS-CoV-2. Using the Illumina COVIDSeq Test, Aegis sequenced 2000 to 18,000 samples per week. At one point, the company was responsible for about 15% of all North American sequences deposited in the Global Initiative to Share All Influenza Data (GISAID).
“We were hiring and onboarding 50 people a week for eight weeks,” says Matthew Hardison, PhD, senior vice president for Lab Operations at Aegis. “We completed four construction projects to expand capacity without shutting down for one minute.”
Prior to the pandemic, Aegis had one Illumina MiSeq. Once their surveillance efforts ramped up, they quickly bought two NovaSeq 6000 Systems, and Illumina lent them a third to help them keep up. It was the beginning of a beautiful friendship.
“We’ve had a great, collaborative relationship with Illumina,” says Senior Scientist Cyndi Clark, PhD. “They helped us scale quickly, and the support has been incredible. If we needed an engineer or a field application scientist, they were there that day or the next day.”
Beyond SARS-CoV-2 pathogen surveillance
As the COVID crisis abated, Aegis had to repurpose its hard-won infectious disease capabilities. The company saw great value in continued surveillance and launched a more comprehensive testing menu.
They quickly transitioned from COVIDSeq to the Illumina Respiratory Pathogen Infectious Disease/Antimicrobial Resistance Panel (RPIP), which covers more than 280 microbes. The panel can identify and characterize bacteria, viruses, fungi, antibiotic resistance genes, and even novel pathogens. The Aegis team was particularly impressed by Illumina’s Explify RPIP data analysis software on BaseSpace Sequence Hub (BSSH). A recent addition to the Illumina Connected Software portfolio, the RPIP application transforms raw FASTQ files into scalable, actionable insights. It simultaneously detects, profiles, and quantifies DNA and RNA respiratory pathogens on the Amazon Web Services (AWS) cloud.
“We don’t have a bioinformatics specialist on the team, but Illumina has built this end-to-end process where we don’t need one,” says Clark. “We can focus on being experts in infectious diseases for things like distinguishing normal flora, but in terms of making pathogen calls from huge sequencing data files, the software does everything and does it quick.”
“The infectious disease space is moving from culture-based identification to molecular—it’s faster, more sensitive, and provides more information,” says Hardison. “On a public health level, broad pathogen surveillance is incredibly important to respond to COVID or any emerging pathogens.”
Funding from the Bill & Melinda Gates Foundation
Aegis’s capacity and quality put it on the CDC’s radar to provide SARS-CoV-2 variant tracking. Later, the National Institutes of Health contracted with the lab to conduct SARS-CoV-2 genotyping. These collaborations, and the lab’s overall capabilities, led to funding from the Bill & Melinda Gates Foundation.
During the pandemic, clinicians, labs, and public health officials were narrowly focused on SARS-CoV-2—but were other pathogens in the mix? Aegis was uniquely positioned to answer that question. The lab selected 4400 biobank nasal swab samples—previously tested for SARS-CoV-2 only—from all 50 states plus the District of Columbia, Puerto Rico, and the US Virgin Islands. The lab also possessed deidentified symptom, risk factor, and demographic data they could link to the sequencing output.
Aegis received a $2.8 million grant from the Bill & Melinda Gates Foundation to reexamine those samples and identify possible co-infections. RPIP gave the lab the necessary power to perform this retrospective study.
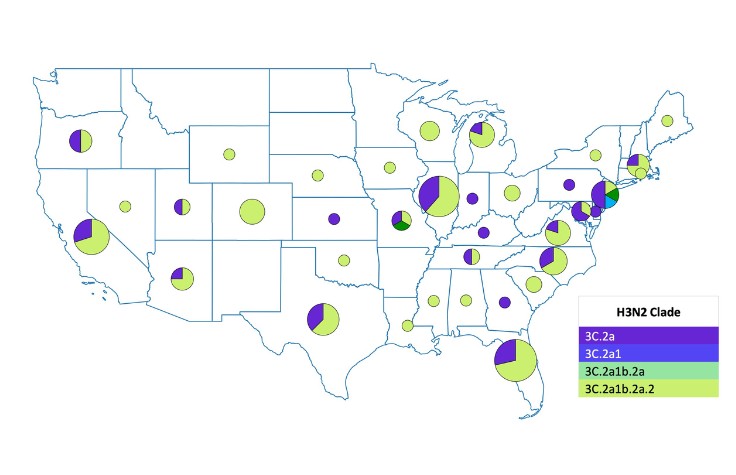
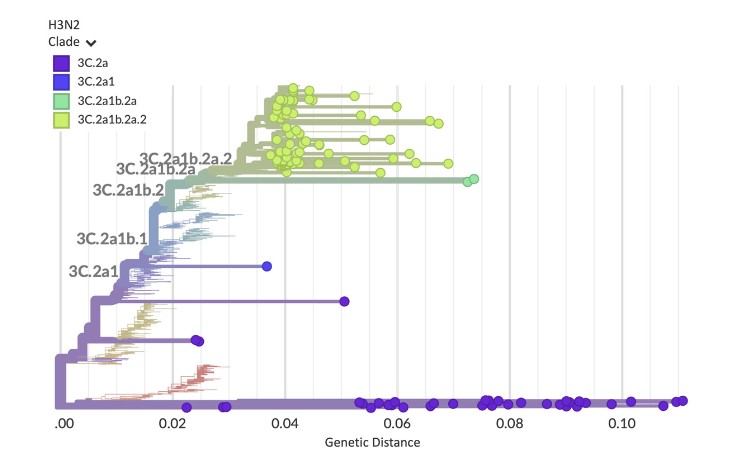
“The underlying goal was to create an approach we could take to a developing country as a guidebook on doing multi-pathogen surveillance at scale and not just looking at one pathogen at a time,” says Hardison. “We created capacity models and cost models to show how to run this workflow to boost turnaround times, increase cost efficiencies, and get it up and running in the lab.”
Aegis continues to collaborate with Illumina on the RPIP data and is pursuing possible publications. The lab also conducted a study with Pfizer to examine vaccine effectiveness, and is working with the University of Wisconsin to compare environmental (air and wastewater) samples with traditionally collected clinical specimens as a proof of concept for early outbreak detection.
These are only a few of the potential studies Aegis and its collaborators can conduct. The ability to rapidly respond, collaborate, and scale the broad assessment of pathogens and host genomes opens a wide world of possible interrogations.
“We are heavily focused on how we can align the newest, best molecular approaches in conjunction with more old school (but still effective) methodologies to determine antibiotic resistance,” says Hardison. “Certainly, antibiotic stewardship is going to become more important in coming years.”


