
Time
To process a typical whole human genome with 30× coverage, a variety of compute platforms can be employed. Some applications demand speedy secondary analysis to get answers quicker. Other applications may benefit from speedy secondary analysis simply because of the volume of data being produced, and NovaSeq X embedded secondary analysis is more than capable of meeting such demand.
Producing up to 128 or more 30× whole-genome sequencing (WGS) samples per dual-flow-cell run, the NovaSeq X is a true factory-scale instrument. But tucked into the sleek shell of the NovaSeq X is a hidden DRAGEN capable of performing secondary analysis on the fly, often processing the data of the last run in only as much time as it takes to set up the next (that is, the wash and clustering time).
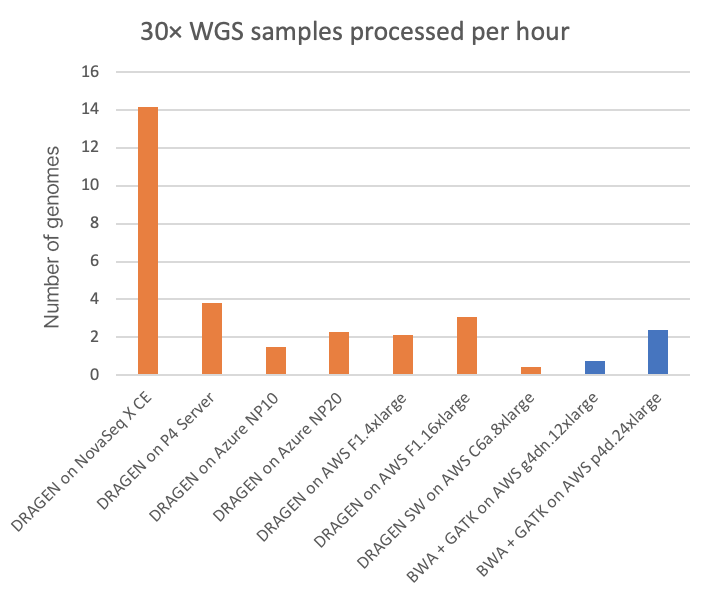
For the NovaSeq X Plus, on-instrument DRAGEN processing is performed on a dual socket AMD EPYC 7552 (with a combined 96 x86_64 cores) that also has at its disposal four Xilinx Alveo U250 FPGA cards and 1.5 TB of RAM. This significant compute horsepower allows the on-instrument DRAGEN to output to gVCF a fully processed (sort/dedup from per-sample BCL, align reads, variant call) 30× WGS samples about once every four minutes. This speed is compared to some other DRAGEN implementations in Figure 2, below, as well as to some GPU-accelerated third-party pipelines.
In Figure 2, the FPGA-accelerated, embedded DRAGEN onboard the NovaSeq X shows nearly 20× or 6× speedup over T4 or A100 GPU cloud instances, respectively.
Note that in the charts that follow:
- “DRAGEN on NovaSeq X CE” is the embedded, under-the-hood HPC compute that is part of the NovaSeq X instrument.
- “DRAGEN on P4 Server” is the current version of the on-site, dedicated DRAGEN server available for purchase from Illumina.
- All other metrics are derived from running on various kinds of publicly accessible cloud compute instances from Microsoft Azure and Amazon Web Services (AWS).
- All DRAGEN runs involve FPGA acceleration except for those pertaining to “DRAGEN SW on AWS C6a.8xlarge,” which uses a software-only version of the DRAGEN pipeline.
- The BWA + GATK runs are shown for two different types of GPU instance types in the AWS cloud, where the pipelines have been ported to—and optimized for—GPU for accelerated runtimes.
Cost
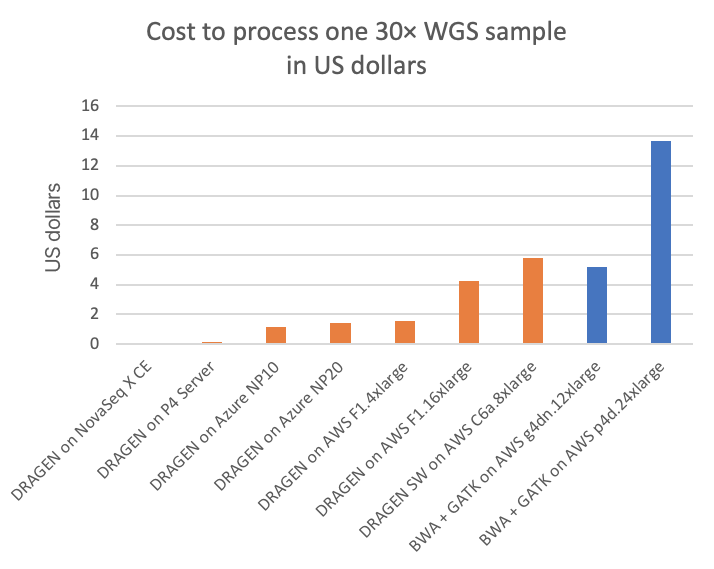
When large amounts of data are being processed, cost of compute must also be considered. In the NovaSeq X instruments, the DRAGEN compute is included as part of the instrument cost and, therefore, is effectively free to NovaSeq X users. However, if we were to amortize the compute component cost over the sequencing throughput and expected five-year life of the instrument, we would arrive at a cost per 30× WGS sample.
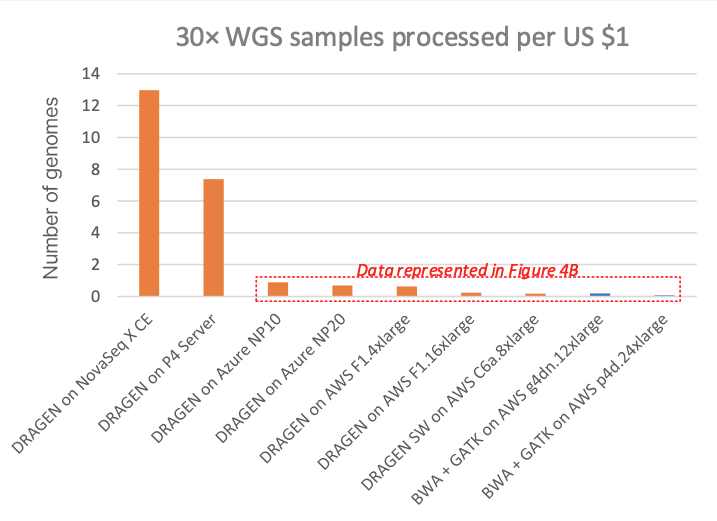
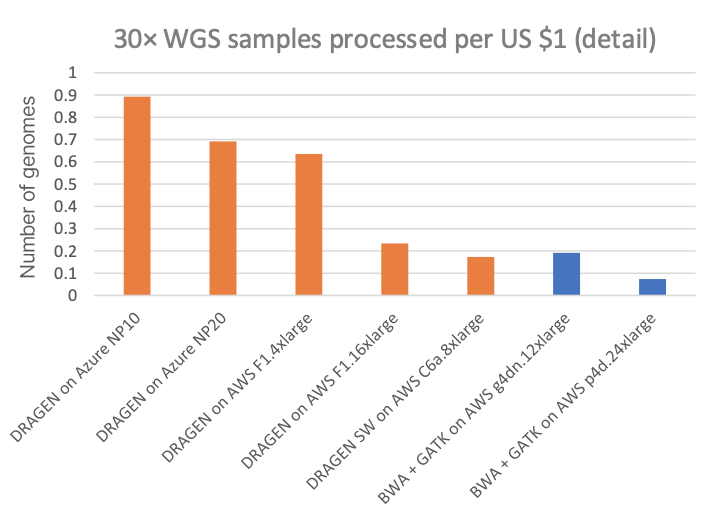
Figure 3 compares this to several other DRAGEN and third-party pipelines on various cloud compute instances (“on-demand” pricing shown for all cloud listings), in cost per 30× WGS sample. Figure 4A shows how many 30× WGS samples can be processed per compute dollar, and subplot Figure 4B shows a detail view of Figure 4A with the dedicated DRAGEN hardware data points removed (the NovaSeq X CE and the DRAGEN P4 Server samples-per-dollar numbers are so high that the nuances in pricing for the cloud options require an expansion of the y-axis scale for better visibility).
In the charts, the FPGA-accelerated DRAGEN onboard the NovaSeq X shows up to more than 60× or 170× cost advantage over T4 or A100 GPU cloud instances, respectively.
Energy
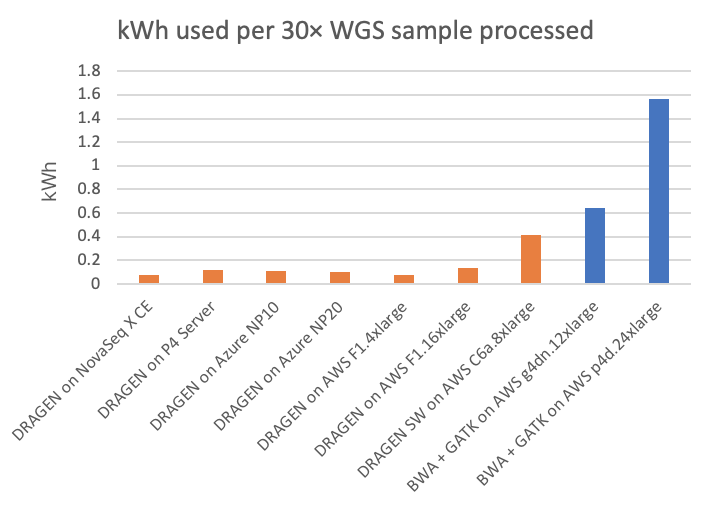
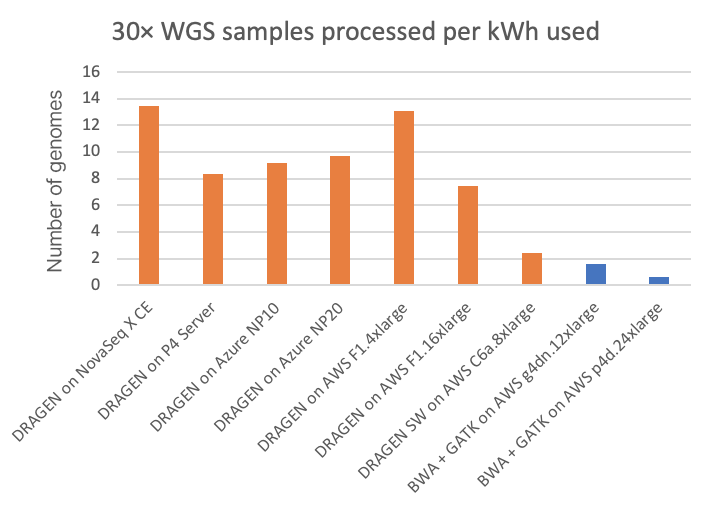
Power consumption is a factor of interest, not only as it relates to environmental impact and utilities costs, but also because power may limit the form factor of the secondary analysis compute platform. On the NovaSeq X CE and P4 Server DRAGEN runs, we can directly measure the power consumption from the DRAGEN compute platform. On the cloud platforms, we are estimating power consumption based on posted system configurations. (Reader, if you have more accurate information on cloud power consumption, please contact us using the form linked here and we will happily amend this article accordingly.) Energy usage, both in kilowatt-hours per 30× WGS sample and its inverse (30× WGS per kWh), is shown in Figure 5 and Figure 6, respectively, for the on-instrument NovaSeq X DRAGEN and on-site P4 DRAGEN server, as well as for other DRAGEN and third-party pipelines running on cloud compute instances.
In the charts, the FPGA-accelerated DRAGEN onboard the NovaSeq X shows up to more than 8× or 20× energy efficiency advantage over T4 or A100 GPU cloud instances, respectively.
Footprint
Notably, the NovaSeq X instrument has only a single standard 200–240 volt AC, 50/60 hertz, 15 amp, single-phase power plug, from which both the sequencing hardware (lasers, heaters, pumps, and so on) and the DRAGEN compute draw their power. Additionally, heat generated from the embedded DRAGEN compute must be removable to a degree that it does not adversely affect the chemistry or biology of the sequencing itself. FPGA-accelerated DRAGEN—with its lower energy expenditure per genome and compact HPC-class compute—enables this streamlined footprint.
Quality of results
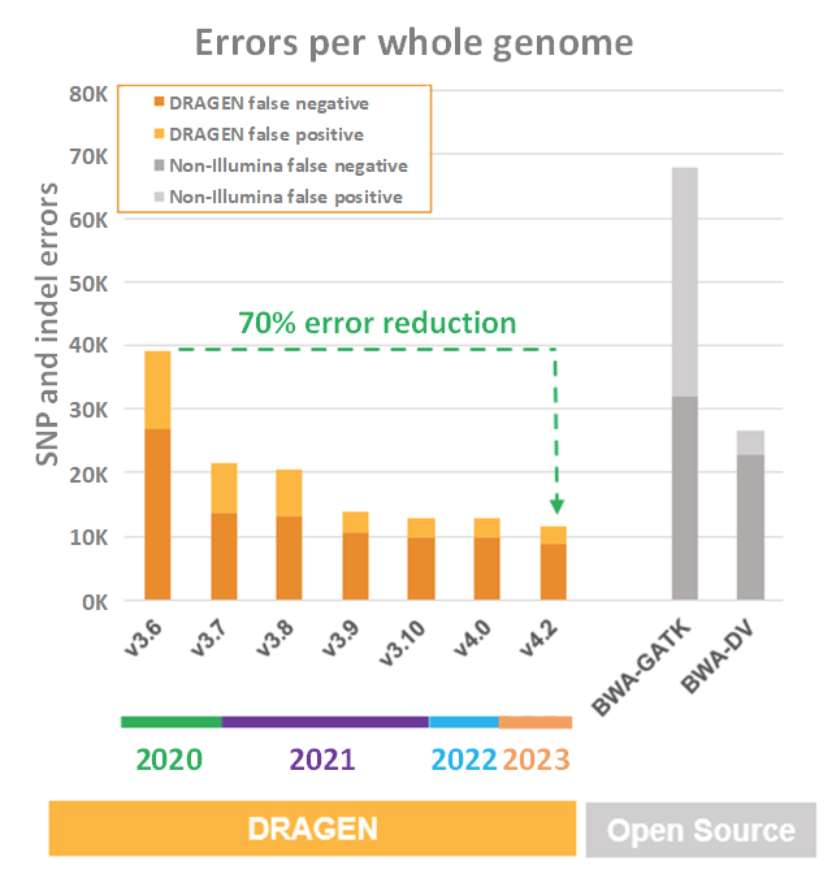
It is impressive enough that the NovaSeq X on-instrument DRAGEN compute comes out on top in all the throughput, cost, power, and footprint comparisons detailed above. But those charts don’t really tell the whole story of why FPGA was chosen to accelerate DRAGEN. FPGA was chosen for DRAGEN acceleration because the superior throughput, cost, power, and footprint features allow the application of more compute-intensive algorithms to the task of wringing as much useful information as possible from the sequencer reads. DRAGEN has become so good at doing this that it is now its own fiercest competition, relentlessly driving up precision, sensitivity, and specificity with each successive release of the DRAGEN pipeline. This is summarized in Figure 7, below, which shows false positive and false negative metrics for both third-party secondary analysis pipelines (BWA + GATK and Google’s Deep Variant) and several generations of DRAGEN.
Conclusion
Both CPU and GPU manufacturers continue to improve their product offerings as they relate to genomic workloads, and programming pipelines in CPU or GPU requires less developer effort than would be required for FPGA.
Nonetheless, Illumina engineers have done the hard work of porting genomic pipelines to FPGA-accelerated platforms, and this FPGA acceleration continues to show significant advantages in every metric that our customers care about. This, in turn, translates to real benefits to those Illumina customers who can use the FPGA-accelerated DRAGEN pipelines.
Much has been said in the industry about the impending compute cliff that genomics faces, as if compute costs, power, and/or turnaround time will somehow bring an abrupt slowing or stop to the advances now being witnessed in genomics. With FPGA acceleration, no such cliff appears to be in sight. On-instrument FPGA-accelerated DRAGEN can fully analyze a 30× human genome in less than five minutes, at a compute cost of less than ten US cents, with about the same energy it takes to propel the most efficient Tesla Model 3 half a kilometer down the road.
The second-to-none throughput, cost, power, and footprint metrics of DRAGEN are impressive on their own. Even more impressive is that DRAGEN achieves these metrics while at the same time producing ever-improving quality of results (precision, sensitivity, specificity) that are better than any other NGS secondary analysis pipeline. As CPUs, GPUs and FPGAs evolves, DRAGEN team is on the hunt for the latest technology for better and more accurate results for our users.
To read more about Illumina DRAGEN Secondary Analysis and to request pricing, click here.
To read more about DRAGEN onboard the NovaSeq X Series, click here.