Hematologic Malignancy Profiling with NGS
Molecular Profiling of Blood Cancers
Hematologic malignancies account for 9.5% of new cancer cases diagnosed in the United States each year.1 The many stages of hematopoietic differentiation provide multiple opportunities for mutations that lead to distinct tumor subtypes.2 Genomic characterization of malignant cells enables hematology researchers to gain insights into disease etiology and how cancer subtypes relate to therapeutic options.
In contrast to traditional single-gene methods, next-generation sequencing (NGS) offers significant advancements in sensitivity and scale, and provides greater visibility into important drivers of hematologic malignancies. NGS panels that target myeloma, lymphoma, and leukemia-associated genes enable rapid, accurate molecular profiling studies.
Targeted NGS Panels for Hematologic Malignancies
Among the broad spectrum of NGS methods, targeted sequencing offers easy and cost-effective entry into cancer genomics research. Targeted NGS panels that focus on myeloid leukemia, lymphoma, and/or other hematologic malignancy-associated genes generate a smaller, more manageable data set than broader methods such as whole-genome or exome sequencing.
Hematologic malignancy profiling with targeted NGS follows a rapid, simple workflow that researchers can easily scale to analyze hundreds of samples. Many samples can be pooled together and sequenced simultaneously, simplifying lab validation procedures and significantly increasing sample throughput.
NGS for Myelodysplastic Syndrome Studies
Learn how NGS complements traditional cytogenetic methods for research into myelodysplastic disorders and other challenging neoplasms.
Featured Hematologic Cancer Research
NGS Panels Enable Pediatric Leukemia Research
A targeted RNA sequencing pan-cancer panel gives clinical researchers insight into the role of fusion genes in pediatric leukemia.
Read Interview
Benefits of a Myeloid Leukemia NGS Panel
Researchers discuss how their targeted NGS panel identifies myeloid disease-related gene mutations with higher sensitivity than Sanger sequencing or cytogenetic methods.
Read Interview
Comprehensive Gene Fusion Detection
Scientists describe the benefits of an NGS-based gene fusion panel for research into hematologic malignancies and other cancer types.
Read ArticleNGS Provides Insights into Leukemia and Lymphoma
Dr Torsten Haferlach of Munich Leukemia Laboratory discusses research plans to sequence 5,000 samples from individuals with different kinds of leukemias and lymphomas. Their goal is to gain a better understanding of leukemia and lymphoma subtypes and discover potential new therapeutic pathways.
Products for Hematologic Malignancy Research
Expert-defined NGS panels for myeloid malignancies, lymphomas, and other hematologic disorders enable rapid analysis of cancer-associated genes. These panels deliver streamlined workflows that enable different sample types to be assessed with the same assay.
Illumina sequencers deliver industry-leading accuracy, generating ~ 90% of the world’s sequencing data.* Efficient reporting capabilities enable researchers to annotate, classify, and communicate significant findings in a standard format that facilitates data analyses and trending.
Click on the below to view products for each workflow step.
Targeted panel to investigate 40 DNA genes, 29 RNA fusion driver genes, and 5 gene expression levels associated with myeloid malignancies and disorders.
TruSight RNA FusionA gene fusion detection panel targeting fusion associated genes in many cancer types with the ability to detect known and novel fusion gene partners.
TruSight RNA Pan-CancerTargeting 1385 cancer-associated genes for gene expression, variant and fusion detection in all cancer research sample types including FFPE.
A highly multiplexed polymerase chain reaction (PCR)-based workflow for use with targets ranging from a few to hundreds of genes in a single run.
TruSight Myeloid Sequencing PanelExpert-defined DNA analysis panel targeting oncogenic exons and tumor suppressor genes associated with myeloid cancers.
TruSight Oncology UMI ReagentsThe TruSight Oncology UMI Reagents and UMI Error Correction App reduce error rates in samples to ≤0.007%, enabling the detection of low frequency variants.
Small and affordable benchtop sequencer for reliable low-throughput targeted sequencing of hematological cancers.
MiSeq SystemDesktop sequencer featuring a simple NGS workflow, integrated data analysis software, and unmatched accuracy.
FDA-cleared in vitro diagnostic NGS system that also operates in research mode to generate accurate, reliable data.
NextSeq 550 SystemFlexible desktop sequencer that supports multiple applications, from targeted profiling to whole-genome sequencing.
The Illumina genomics computing environment for NGS data analysis and management.
Local Run ManagerEasy-to-use software for automated on-instrument data analysis.
A powerful analysis and reporting tool that provides biological insight into genomic variant data.
MiSeq ReporterEasy-to-use software for analysis and variant calling on the MiSeq System.
Webinar: Genetic and Genomic Approaches to Modeling Clonal Evolution in Myeloid Malignancies
In this informative webinar, Bobby Bowman, PhD walks through approaches in modeling clonal evolution in myeloid malignancies. He discusses the use of a custom amplicon panel to delineate co-occurring mutations at a clonal level, querying the 31 most recurrently mutated genes in myeloid disease, and more. These studies provide new insights into the stages of malignant transformation, clonal diversity, and oncogene-differentiate state relationships.
Watch Webinar
Learn More
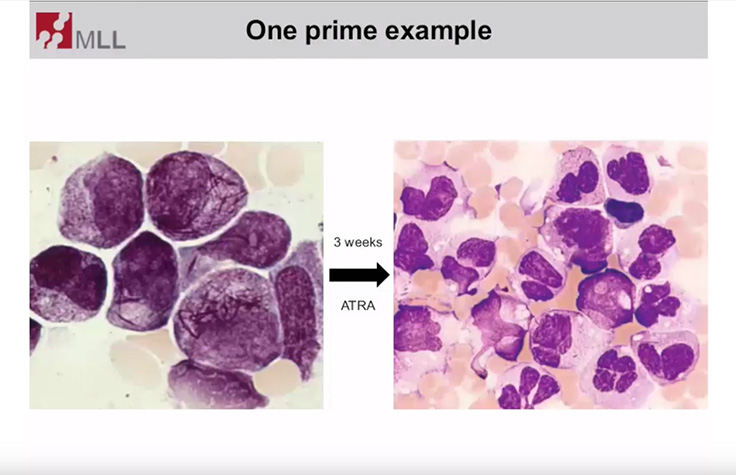
Molecular Assessment of Hematologic Disorders
Learn more about the benefits of targeted sequencing panels for researching hematologic malignancies.
Targeted Resequencing Analysis for Myeloid Disorders
Researchers used NGS panels to analyze 25 genes frequently mutated in myeloid malignancies.
Interested in receiving newsletters, case studies, and information on cancer genomics?
Sign UpReferences
- Leukemia & Lymphoma Society (www.lls.org)
- Staudt LM. Molecular diagnosis of the hematologic cancers. N Engl J Med. 2003;348:1777-1785.
* Data calculations on file. Illumina, Inc. 2015.
