Advantages of cfRNA for noninvasive health monitoring
Circulating cell-free RNA (cfRNA; also called C-RNA) is a promising alternative for noninvasive assessment of health. Unlike DNA, a cell's RNA transcriptome is dynamic and tissue specific. cfRNA is released by various tissues into circulation via the cellular processes of apoptosis, microvesicle shedding, and exosome signaling. cfRNA is also stable, contained within vesicles that protect it from degradation by nucleases. Because of its diverse origins, cfRNA measurements reflect tissue-specific changes in gene expression, intercellular signaling, and the degree of cell death occurring within different tissues throughout the body (Figure 1).5-9
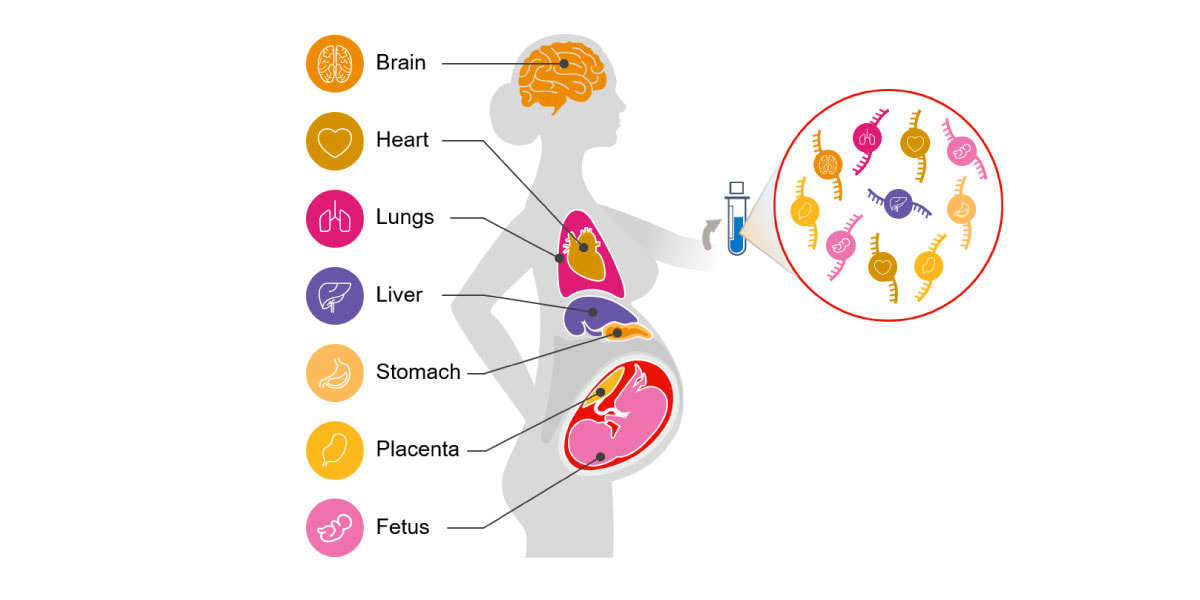
Figure 1. Circulating cfRNA can be obtained from a standard blood draw and reflects molecular changes in all body systems.
Optimized protocol for cfRNA library preparation and sequencing
While cfRNA has high informative potential, it does have practical challenges. Each milliliter of plasma yields only a few nanograms of cfRNA, which is a mixture of full-length RNA and fragmented RNA. The most abundant transcripts present in human blood are ribosomal RNA (rRNA) and globulin RNA, which can overpower the signal of more informative transcripts. To address these issues, our research team at Illumina developed a workflow that was amenable to the low yield and partially fragmented nature of cfRNA. Rather than use standard depletion methods to remove rRNA, we generated a library from all cfRNA and carried out probe-assisted enrichment targeting the whole human exome.
The optimized workflow for cfRNA sequencing maximizes exonic cfRNA signal, minimizes risk failure for low-input samples, and is compatible with overnight shipment of blood samples at room temperature. The original protocol used ligation-based library preparation to generate high-quality whole-transcriptome cfRNA sequencing data.1 Further development efforts showed that tagmentation-based RNA library prep kits enable even greater sensitivity and throughput for the next wave of research (Figure 2, Figure 3).10
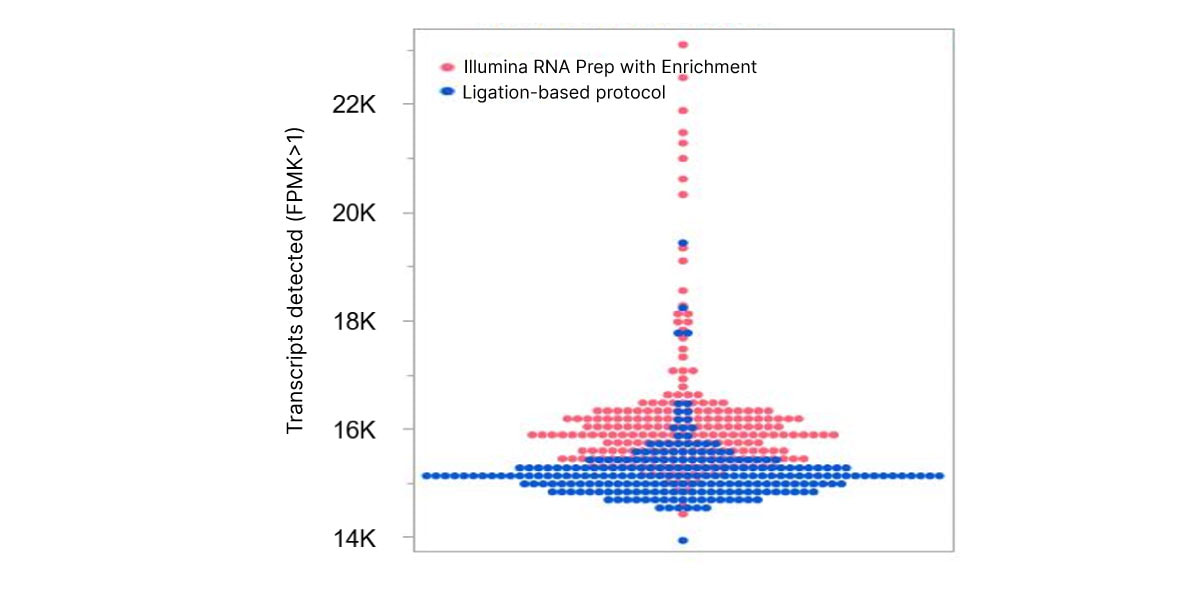
Figure 2. Illumina RNA Prep with Enrichment enables greater sensitivity to detect more circulating transcripts than with the ligation-based protocol.
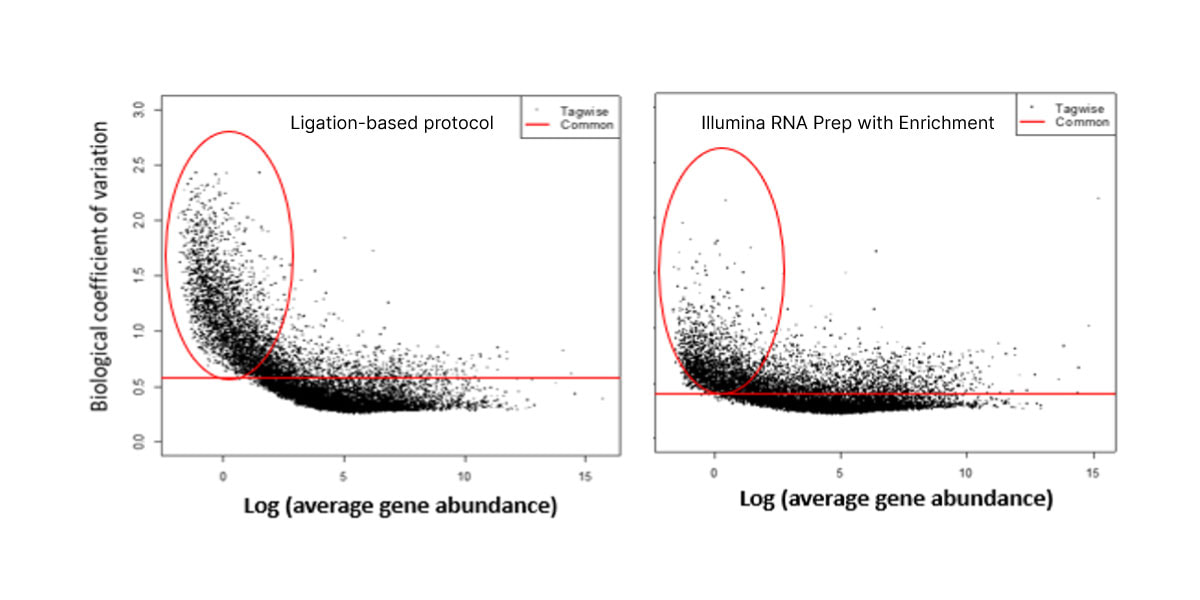
Figure 3. Illumina RNA Prep with Enrichment shows decreased noise for low-abundance cfRNA transcripts than with the ligation-based protocol.
Proof-of-concept study to identify cfRNA signatures in early-onset preeclampsia
Our recent Science Translational Medicine publication described how we established and applied this workflow in a proof-of-concept study to identify cfRNA alterations associated with the dangerous pregnancy complication preeclampsia.1 The team collected 113 blood samples, including 40 from women with severe, early-onset preeclampsia and 73 from women with healthy pregnancies.
Aided by machine learning, adapted to clarify trends in sometimes noisy data, we discovered a cfRNA signature that captures the dysfunction of diverse body systems and accurately classifies preeclampsia status across independent cohorts. We identified 30 transcripts in circulation that were differentially abundant compared with noncomplicated pregnancies. These transcripts had gene ontology annotations and tissue expression patterns consistent with the placental dysfunction, impaired fetal development, and maternal immune and cardiovascular dysregulation characteristic of preeclampsia. (Table 1).
Highlights of identified circulating transcripts associated with preeclampsia
The cfRNA changes we observed in preeclampsia patients matched the biology of the disease and corroborated the validity of this approach. Of the 30 identified gene transcripts, over half were previously associated with preeclampsia (Table 1).11-29 The altered cfRNA transcripts captured information from multiple different organ systems, representing contributions from maternal, placental, and fetal tissues. For instance, we identified dysfunction in the placenta, as well as altered blood pressure regulation in the mother.
The following gene categories highlight specific examples of the impaired biological processes captured by cfRNA.
IGF signaling: impaired fetal growth and development
Preeclampsia is associated with low levels of bioavailable insulin-like growth factor (IGF), which is a major regulator of fetal growth and development.30-32 IGFBP5, PAPPA2, HTRA4, and PRG2 are all relevant to the regulation of IGF signaling and were found to be increased in early-onset preeclampsia subjects.
Pregnancy duration: risk of preterm delivery
CRH, ZEB1, and PNMT, genes associated with pregnancy duration and the onset of labor,33-35 were upregulated in preeclamptic cfRNA. Preterm delivery is a common outcome of early-onset preeclampsia and nearly all affected participants in this study delivered prematurely.
Angiogenesis: compromised placental development
Ten transcripts identified in this study are important for angiogenesis regulation: APOLD1, LEP, SEMA3G, ADAMTS1, TIMP3, ADAMTS2, HTRA4, HSPA12B, SLC9A3R2, and TIMP4. Angiogenesis is a crucial component of proper placental development throughout gestation, but is impaired in preeclamptic pregnancies.36,37
Blood pressure regulation: hypertension
Hypertension is a defining symptom of preeclampsia. Multiple transcripts associated with blood pressure regulation, ALOX15B, PNMT, ARHGEF25, APOLD1, and CRH, showed elevated levels of cfRNA transcripts in this cohort.
Dendritic cell activity: impaired pregnancy immune tolerance
Immune system dendritic cell markers also exhibited anomalous expression in preeclampsia patients: CLEC4C, PLD4, TIMP3, and VSIG4. Dendritic cells play a key role in the immune system’s tolerance of the placenta and fetus in pregnancy and have been implicated as a potential driving factor behind preeclampsia.38-40
| Gene symbol | Fold change in preeclampsia | Protein function |
|---|---|---|
Fetal tissue expression |
||
IGFBP5* |
+3.6 |
IGF signaling |
ALOX15B* |
+5.7 |
Blood pressure regulation |
NES* |
+4.5 |
Fetal development |
TEAD4 |
+3.3 |
Cell proliferation |
PITPNM3 |
+3.2 |
Cell proliferation |
CUX2 |
-3.3 |
Cell proliferation |
FAM107A |
+5.0 |
Cell proliferation and migration |
PRX* |
+3.8 |
Cell structure |
AMPH |
+5.0 |
Endocytosis |
Placental tissue expression |
||
PAPPA2* |
+4.9 |
IGF signaling |
HTRA4* |
+3.9 |
IGF signaling; angiogenesis |
LEP* |
+10.7 |
Angiogenesis |
SEMA3G |
+3.5 |
Angiogenesis |
APOLD1* |
+3.4 |
Angiogenesis; blood pressure regulation |
VSIG4* |
+8.1 |
Dendritic cell activity |
Placental/fetal tissue expression |
||
PRG2* |
+5.2 |
IGF signaling |
CRH* |
+5.7 |
Pregnancy duration; blood pressure regulation |
ADAMTS1* |
+3.5 |
Angiogenesis |
ADAMTS2 |
+11.6 |
Angiogenesis |
TIMP3* |
+4.1 |
Angiogenesis; dendritic cell activity |
Other tissue expression |
||
PNMT |
+3.8 |
Pregnancy duration; blood pressure regulation |
ZEB1* |
+2.8 |
Pregnancy duration; blood pressure regulation |
TIMP4* |
+4.3 |
Angiogenesis |
SLC9A3R2 |
+3.6 |
Angiogenesis |
HSPA12B |
+3.5 |
Angiogenesis |
ARHGEF25 |
+4.1 |
Blood pressure regulation |
PLD4 |
-3.0 |
Dendritic cell activity |
CLEC4C* |
-3.6 |
Dendritic cell activity |
DAAM2* |
+5.6 |
Cell proliferation |
KRT5* |
-5.8 |
Cell structure |
* Genes previously associated with preeclampsia11-29
Noninvasive biomarker discovery and diagnostics for complex disorders
Our findings illustrate how sequencing cfRNA can detect molecular signals of preeclampsia in the blood, monitoring maternal, fetal, and placental functions in real time. This study was a proof-of-concept to show the presence of a stable disease signature in the blood. Samples were collected from patients at time of diagnosis, meaning symptoms were already manifesting. However, cfRNA transcripts indicative of preeclampsia could be present prior to symptom onset and further research is needed to characterize the potential for predictive biomarker discovery. This detection model and screening for preeclampsia in the first trimester of pregnancy could potentially identify women at increased risk of developing pregnancy complications and allow for early intervention.41
This work highlights the broad potential of cfRNA as a research and biomarker discovery tool for noninvasive, comprehensive molecular monitoring of health. The optimized protocol for cfRNA sequencing enables robust detection of circulating transcripts to identify reproducible, biologically relevant changes associated with disease. More researchers are evaluating the utility of circulating RNA signatures as accessible and unique biomarkers for diagnosis and surveillance of Alzheimer’s disease, cancer, and other complex diseases.42-46 Further application of this technology offers the possibility to transform our understanding, diagnosis, and treatment of many conditions.
Learn More
Application note: Improved detection of circulating transcripts
Product page: Illumina RNA Prep with Enrichment
References
- Munchel S, Rohrback S, Randise-Hinchliff C, et al. Circulating transcripts in maternal blood reflect a molecular signature of early-onset preeclampsia. Sci Transl Med.2020;12(550):eaaz0131. doi:10.1126/scitranslmed.aaz0131
- Parikh AR, Leshchiner I, Elagina L, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med.2019;25(9):1415-1421. doi:10.1038/s41591-019-0561-9
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non small Cell Lung Cancer. Clin Cancer Res.2019;25(15):4691 4700. doi:10.1158/1078 0432.CCR 19 062z
- Palmero R, Taus A, Viteri S, et al. Biomarker Discovery and Outcomes for Comprehensive Cell Free Circulating Tumor DNA Versus Standdard of Care Tissue Testing in Advanced Non Small Cell Lung Cancer. JCO Precision Oncology.2021;5:93 102. doi:10.1200/PO.20.00241
- Turchinovich A, Drapkina O, Tonevitsky A. Transcriptome of extracellular vesicles: State-of-the-art. Front Immunol.2019;10:202. doi:10.3389/fimmu.2019.00202
- Anfossi S, Babayan A, Pantel K, Calin G. Clinical utility of circulating noncoding RNAs—an update. Nat Rev Clin Oncol.2018;15:541-563.
- Li Y, Elashoff D, Oh M, et al. Serum circulating human mRNA profiling and its utility for oral cancer detection. J Clin Oncol.2006;24:1754-1760. doi:10.1200/JCO.2005.03.7598
- Starling S. A molecular signal for preeclampsia. Nat Rev Endocrinol.2020;16:471. doi:10.1038/s41574-020-0397-x
- Lo Y, Ng K, Tsui B, Chiu W, inventors; Chinese University of Hong Kong, assignee. Circulating mRNA as diagnostic markers. US patent application 20040203037A1. October 14, 2004.
- Illumina. Improved detection of circulating transcripts. Accessed November 19, 2021.
- Jia Y, Li T, Huang X, et al. Dysregulated DNA Methyltransferase 3A Upregulates IGFBP5 to Suppress Trophoblast Cell Migration and Invasion in Preeclampsia. Hypertension.2017;69(2):356-366. doi:10.1161/HYPERTENSIONAHA.116.08483
- Wang Y, Zhu D, An Y, Sun J, Cai L, Zheng J. Preeclampsia activates 15-lipoxygenase and its metabolite 15-hydroxyeicosatetraenoic acid enhances constriction in umbilical arteries. Prostaglandins Leukot Essent Fatty Acids.2012;86(1-2):79-84. doi:10.1016/j.plefa.2011.10.006
- Hwang HS, Cho NH, Maeng YS, Kang MH, Park YW, Kim YH. Differential expression of nestin in normal and pre-eclamptic human placentas. Acta Obstet Gynecol Scand.2007;86(8):909-914. doi:10.1080/00016340701417018
- Tan KH, Tan SS, Sze SK, Lee WK, Ng MJ, Lim SK. Plasma biomarker discovery in preeclampsia using a novel differential isolation technology for circulating extracellular vesicles. Am J Obstet Gynecol.2014;211(4):380.e1-380.e3813. doi:10.1016/j.ajog.2014.03.038
- Kramer AW, Lamale-Smith LM, Winn VD. Differential expression of human placental PAPP-A2 over gestation and in preeclampsia. Placenta.2016;37:19-25. doi:10.1016/j.placenta.2015.11.004
- Singh H, Zhao M, Chen Q, et al. Human HtrA4 Expression Is Restricted to the Placenta, Is Significantly Up-Regulated in Early-Onset Preeclampsia, and High Levels of HtrA4 Cause Endothelial Dysfunction. J Clin Endocrinol Metab.2015;100(7):E936-E945. doi:10.1210/jc.2014-3969
- Xiang Y, Cheng Y, Li X, et al. Up-regulated expression and aberrant DNA methylation of LEP and SH3PXD2A in pre-eclampsia. PLoS One.2013;8(3):e59753. doi:10.1371/journal.pone.0059753
- Junus K, Centlow M, Wikström AK, Larsson I, Hansson SR, Olovsson M. Gene expression profiling of placentae from women with early- and late-onset pre-eclampsia: down-regulation of the angiogenesis-related genes ACVRL1 and EGFL7 in early-onset disease. Mol Hum Reprod.2012;18(3):146-155. doi:10.1093/molehr/gar067
- Textoris J, Ivorra D, Ben Amara A, et al. Evaluation of current and new biomarkers in severe preeclampsia: a microarray approach reveals the VSIG4 gene as a potential blood biomarker. PLoS One.2013;8(12):e82638. doi:10.1371/journal.pone.0082638
- Gormley M, Ona K, Kapidzic M, Garrido-Gomez T, Zdravkovic T, Fisher SJ. Preeclampsia: novel insights from global RNA profiling of trophoblast subpopulations. Am J Obstet Gynecol.2017;217(2):200.e1-200.e17. doi:10.1016/j.ajog.2017.03.017
- Ng EK, Leung TN, Tsui NB, et al. The concentration of circulating corticotropin-releasing hormone mRNA in maternal plasma is increased in preeclampsia. Clin Chem.2003;49(5):727-731. doi:10.1373/49.5.727
- Purwosunu Y, Sekizawa A, Farina A, et al. Cell-free mRNA concentrations of CRH, PLAC1, and selectin-P are increased in the plasma of pregnant women with preeclampsia. Prenat Diagn.2007;27(8):772-777. doi:10.1002/pd.1780
- Namlı Kalem M, Kalem Z, Yüce T, Soylemez F. ADAMTS 1, 4, 12, and 13 levels in maternal blood, cord blood, and placenta in preeclampsia. Hypertens Pregnancy.2018;37(1):9-17. doi:10.1080/10641955.2017.1397690
- Xie D, Zhu J, Liu Q, et al. Dysregulation of HDAC9 Represses Trophoblast Cell Migration and Invasion Through TIMP3 Activation in Preeclampsia. Am J Hypertens.2019;32(5):515-523. doi:10.1093/ajh/hpz006
- Yang X, Meng T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene.2019;683:225-232. doi:10.1016/j.gene.2018.10.015
- Sandrim VC, Diniz S, Eleuterio NM, Gomes KB, Dusse LMS, Cavalli RC. Higher levels of circulating TIMP-4 in preeclampsia is strongly associated with clinical parameters and microRNA. Clin Exp Hypertens.2018;40(7):609-612. doi:10.1080/10641963.2017.1411499
- Darmochwal-Kolarz D, Rolinski J, Tabarkiewicz J, et al. Myeloid and lymphoid dendritic cells in normal pregnancy and pre-eclampsia. Clin Exp Immunol.2003;132(2):339-344. doi:10.1046/j.1365-2249.2003.02136.x
- Løset M, Mundal SB, Johnson MP, et al. A transcriptional profile of the decidua in preeclampsia. Am J Obstet Gynecol.2011;204(1):84.e1-84.e27. doi:10.1016/j.ajog.2010.08.043
- Tan KH, Tan SS, Sze SK, Lee WK, Ng MJ, Lim SK. Plasma biomarker discovery in preeclampsia using a novel differential isolation technology for circulating extracellular vesicles. Am J Obstet Gynecol.2014;211(4):380.e1-380.e3813. doi:10.1016/j.ajog.2014.03.038
- Vatten LJ, Nilsen TI, Juul A, Jeansson S, Jenum PA, Eskild A. Changes in circulating level of IGF-I and IGF-binding protein-1 from the first to second trimester as predictors of preeclampsia. Eur J Endocrinol.2008;158(1):101-105. doi:10.1530/EJE-07-0386
- Halhali A, Tovar AR, Torres N, Bourges H, Garabedian M, Larrea F. Preeclampsia is associated with low circulating levels of insulin-like growth factor I and 1,25-dihydroxyvitamin D in maternal and umbilical cord compartments. J Clin Endocrinol Metab.2000;85(5):1828-1833. doi:10.1210/jcem.85.5.6528
- Kappou D, Vrachnis N, Sifakis S. Recent Insights into the Role of the Insulin-Like Growth Factor Axis in Preeclampsia. In: Sifakis S, ed. From Preconception to Postpartum.InTech; 2012:147-160.
- Xu YJ, Ren LD, Zhai SS, et al. OXTR and ZEB1 expression before and after progesterone dosing in pregnant women with threatened premature labor. Eur Rev Med Pharmacol Sci.2017;21(14):3164-3168.
- Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A.2010;107(48):20828-20833. doi:10.1073/pnas.1008301107
- Smith R, Nicholson RC. Corticotrophin releasing hormone and the timing of birth. Front Biosci.2007;12:912-918. doi:10.2741/2113
- Cerdeira AS, Karumanchi SA. Angiogenic factors in preeclampsia and related disorders. Cold Spring Harb Perspect Med.2012;2(11):a006585. doi:10.1101/cshperspect.a006585
- Maynard SE, Karumanchi SA. Angiogenic factors and preeclampsia. Semin Nephrol.2011;31(1):33-46. doi:10.1016/j.semnephrol.2010.10.004
- Li J, Huang L, Wang S, Zhang Z. The prevalence of regulatory T and dendritic cells is altered in peripheral blood of women with pre-eclampsia. Pregnancy Hypertens.2019;17:233-240. doi:10.1016/j.preghy.2019.07.003
- Huang SJ, Chen CP, Schatz F, Rahman M, Abrahams VM, Lockwood CJ. Pre-eclampsia is associated with dendritic cell recruitment into the uterine decidua. J Pathol.2008;214(3):328-336. doi:10.1002/path.2257
- Wang J, Tao YM, Cheng XY, et al. Dendritic cells derived from preeclampsia patients influence Th1/Th17 cell differentiation in vitro. Int J Clin Exp Med.2014;7(12):5303-5309.
- Rasmussen M, Reddy M, Nolan R, et al. RNA profiles reveal signatures of future health and disease in pregnancy. Nature.2022;10.1038/s41586-021-04249-w. doi:10.1038/s41586-021-04249-w
- Metzenmacher M, Váraljai R, Hegedüs B, et al. Plasma Next Generation Sequencing and Droplet Digital-qPCR-Based Quantification of Circulating Cell-Free RNA for Noninvasive Early Detection of Cancer. Cancers (Basel).2020;12(2):353. doi:10.3390/cancers12020353
- Ibarra A, Zhuang J, Zhao Y, et al. Non-invasive characterization of human bone marrow stimulation and reconstitution by cell-free messenger RNA sequencing. Nat Commun.2020;11(1):400. doi:10.1038/s41467-019-14253-4
- Toden S, Zhuang J, Acosta AD, et al. Noninvasive characterization of Alzheimer's disease by circulating, cell-free messenger RNA next-generation sequencing. Sci Adv.2020;6(50):eabb1654. doi:10.1126/sciadv.abb1654
- Hulstaert E, Morlion A, Avila Cobos F, et al. Charting Extracellular Transcriptomes in The Human Biofluid RNA Atlas. Cell Rep.2020;33(13):108552. doi:10.1016/j.celrep.2020.108552
- Larson MH, Pan W, Kim HJ, et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat Commun.2021;12(1):2357. doi:10.1038/s41467-021-22444-1