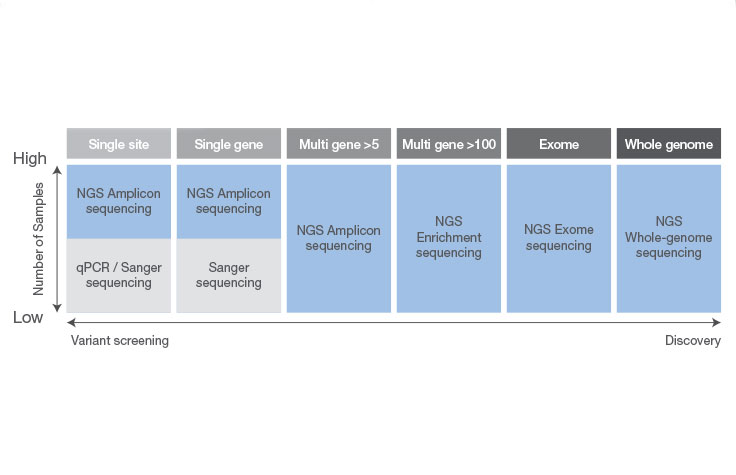
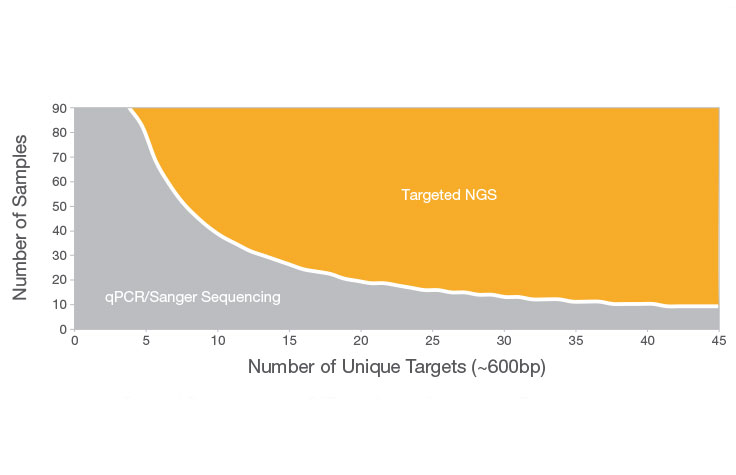
Differences Between NGS and Sanger Sequencing
In principle, the concepts behind Sanger vs. next-generation sequencing (NGS) technologies are similar. In both NGS and Sanger sequencing (also known as dideoxy or capillary electrophoresis sequencing), DNA polymerase adds fluorescent nucleotides one by one onto a growing DNA template strand. Each incorporated nucleotide is identified by its fluorescent tag.
The critical difference between Sanger sequencing and NGS is sequencing volume. While the Sanger method only sequences a single DNA fragment at a time, NGS is massively parallel, sequencing millions of fragments simultaneously per run. This process translates into sequencing hundreds to thousands of genes at one time. NGS also offers greater discovery power to detect novel or rare variants with deep sequencing.

Benefits of NGS vs. Sanger Sequencing
Advantages of NGS include:
- Higher sensitivity to detect low-frequency variants1,2
- Faster turnaround time for high sample volumes3
- Comprehensive genomic coverage
- Lower limit of detection4,5
- Higher capacity with sample multiplexing
- Ability to sequence hundreds to thousands of genes or gene regions simultaneously
Choosing NGS vs. Sanger Sequencing
Explore the benefits and limitations of each method to understand which one best suits your needs.
“With Sanger sequencing, we saw a limited DNA snapshot… NGS and its massively parallel sequencing enable us to look at tens to hundreds of thousands of reads per sample.”
Michael Bunce, PhD
Professor, Head of TrEnD laboratory, Curtin University
Comparison of Sanger Sequencing and NGS
| Sanger Sequencing | Targeted NGS | |
|---|---|---|
| Benefits |
|
|
| Challenges |
|
|
* Discovery power is the ability to identify novel variants.
† Mutation resolution is the size of the mutation identified. NGS can identify large chromosomal rearrangements down to single nucleotide variants.
‡ 10 ng DNA will produce ~1 kb with Sanger sequencing or ~300 kb with targeted resequencing (250 bp amplicon length × 1536 amplicons with an AmpliSeq for Illumina workflow)
Advantages of NGS Over Sanger Sequencing
See examples of recent studies that used NGS to overcome the limitations of Sanger sequencing.
Efficient Variant Discovery with Targeted Gene Panels
NGS enabled Franco Taroni, MD to identify variants in a fraction of the time and at a significantly lower cost than Sanger sequencing.
Read Interview
NGS Revolutionizes Reproductive Genomics
Viafet uses the VeriSeq PGS Solution, enabling IVF clinics to provide fast, accurate, and efficient PGS services.
Read InterviewWhen to Use NGS vs. Sanger Sequencing
Sanger sequencing can be a good choice when interrogating a small region of DNA on a limited number of samples or genomic targets (~20 or fewer). Otherwise, targeted NGS is more likely to suit your needs. NGS allows you to screen more samples cost-effectively and detect multiple variants across targeted areas of the genome—an approach that would be costly and time-consuming using Sanger sequencing.
Watch this animation to see how the easy and accessible Illumina NGS technology can complement your Sanger sequencing work.
Transitioning from Sanger Sequencing to NGS
The iSeq 100 Sequencing System makes it easier and more affordable than ever to bring the power of NGS to your lab. The following example illustrates how the instrument fits into a common NGS workflow.
Library Prep
iSeq-Compatible Library Prep Products
Find the right library prep kit for your sample type and application.
Sequencing
iSeq 100 System
Affordable, fast, and accessible sequencing power for targeted or small genome sequencing in any lab.
Data Analysis
Local Run Manager
An on-site software solution for creating sequencing runs, monitoring run status, and analyzing data.
Featured NGS Methods
Targeted Resequencing

This method involves isolating and sequencing a subset of genes or a genomic region of interest, which can conserve lab resources.
Whole-Genome Sequencing

This method delivers a comprehensive view of genetic variation, ideal for discovery applications.
Related Content
- Advantages of High-Throughput Sequencing: Process more samples to improve statistical power, and cost-effectively run emerging data-rich methods, including single-cell and spatial analyses.
- NGS Data Analysis: Find user-friendly tools and tips to smooth the process of analyzing sequencing data, so you can spend more time doing research and less time configuring workflows.
- Multiomics Profiling: Combine genomic data with data from other modalities such as transcriptomics, epigenetics, and proteomics, to better connect genotype to phenotype and fuel discovery of novel drug targets.
In-Depth Guide to Targeted NGS
Learn more about how NGS-based targeted resequencing can help you identify variants in less time and for less money than Sanger sequencing.
NGS vs. Sanger Sequencing
Interested in receiving newsletters, case studies, and information on genomic analysis techniques? Enter your email address.
Additional Resources
References
- Jamuar SS, Lam AT, Kircher M, et al. Somatic mutations in cerebral cortical malformations. N Engl J Med. 2014;371(8):733-743.
- Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent low-frequency variants associated with inflammatory bowel disease. Nat Genet. 2011;43(11):1066-1073.
- König K, Peifer M, Fassunke J, et al. Implementation of amplicon parallel sequencing leads to improvement of diagnosis and therapy of lung cancer patients. J Thorac Oncol. 2015;10(7):1049-1057.
- Shendure J and Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135-1145.
- Schuster SC. Next-generation sequencing transforms today’s biology. Nat Methods. 2008;5(1):16-18.